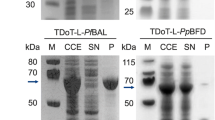
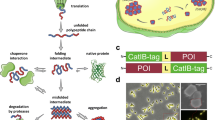
Abstract
Catalytically active inclusion bodies (CatIBs) are promising biologically produced enzyme/protein immobilizates for application in biocatalysis, synthetic chemistry, and biomedicine. CatIB formation is commonly induced by fusion of suitable aggregation-inducing tags to a given target protein. Heterologous production of the fusion protein in turn yields CatIBs. This chapter presents the methodology needed to design, produce, and characterize CatIBs.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Guisan JM (2013) New opportunities for immobilization of enzymes. Methods Mol Biol 1051:1–13
Homaei AA, Sariri R, Vianello F et al (2013) Enzyme immobilization: an update. J Chem Biol 6:185–205
Sheldon RA, Van Pelt S (2013) Enzyme immobilisation in biocatalysis: why, what and how. Chem Soc Rev 42:6223–6235
Ölçücü G, Klaus O, Jaeger KE et al (2021) Emerging solutions for in vivo biocatalysts immobilization: tailor-made catalysts for industrial biocatalysis. ACS Sustain Chem Eng 9:8919–8945
Rehm FB, Chen S, Rehm BH (2016) Enzyme engineering for in situ immobilization. Molecules 21:1370
Schmidt-Dannert S, Zhang G, Johnston T et al (2018) Building a toolbox of protein scaffolds for future immobilization of biocatalysts. Appl Microbiol Biotechnol 102:8373–8388
Steinmann B, Christmann A, Heiseler T et al (2010) In vivo enzyme immobilization by inclusion body display. Appl Environ Microbiol 76:5563–5569
De Marco A, Ferrer-Miralles N, Garcia-Fruitos E et al (2019) Bacterial inclusion bodies are industrially exploitable amyloids. FEMS Microbiol Rev 43:53–72
Garcia-Fruitos E, Gonzalez-Montalban N, Morell M et al (2005) Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb Cell Factories 4:27
Garcia-Fruitos E, Vazquez E, Diez-Gil C et al (2012) Bacterial inclusion bodies: making gold from waste. Trends Biotechnol 30:65–70
Rinas U, Garcia-Fruitos E, Corchero JL et al (2017) Bacterial inclusion bodies: discovering their better half. Trends Biochem Sci 42:726–737
Jäger VD, Lamm R, Küsters K et al (2020) Catalytically-active inclusion bodies for biotechnology-general concepts, optimization, and application. Appl Microbiol Biotechnol 104:7313–7329
Krauss U, Jäger VD, Diener M et al (2017) Catalytically-active inclusion bodies carrier-free protein immobilizates for application in biotechnology and biomedicine. J Biotechnol 258:136–147
Diener M, Kopka B, Pohl M et al (2016) Fusion of a coiled-coil domain facilitates the high-level production of catalytically active enzyme inclusion bodies. ChemCatChem 8:142–152
Jäger VD, Kloss R, Grünberger A et al (2019) Tailoring the properties of (catalytically)-active inclusion bodies. Microb Cell Factories 18:33
Jäger VD, Lamm R, Kloss R et al (2018) A synthetic reaction cascade implemented by colocalization of two proteins within catalytically active inclusion bodies. ACS Synth Biol 7:2282–2295
Kloss R, Karmainski T, Jäger VD et al (2018) Tailor-made catalytically active inclusion bodies for different applications in biocatalysis. Cat Sci Technol 8:5816–5826
Kloss R, Limberg MH, Mackfeld U et al (2018) Catalytically active inclusion bodies of L-lysine decarboxylase from E. coli for 1,5-diaminopentane production. Sci Rep 8:5856
Küsters K, Pohl M, Krauss U et al (2021) Construction and comprehensive characterization of an EcLDCc-CatIB set-varying linkers and aggregation inducing tags. Microb Cell Factories 20:49
Arie JP, Miot M, Sassoon N et al (2006) Formation of active inclusion bodies in the periplasm of Escherichia coli. Mol Microbiol 62:427–437
Huang Z, Zhang C, Chen S et al (2013) Active inclusion bodies of acid phosphatase PhoC: aggregation induced by GFP fusion and activities modulated by linker flexibility. Microb Cell Factories 12:25
Köszagová R, Krajcovic T, Palencarova-Talafova K et al (2018) Magnetization of active inclusion bodies: comparison with centrifugation in repetitive biotransformations. Microb Cell Factories 17:139
Wang R, Li J, Dang D et al (2020) Bacterial production of maize and human serine racemases as partially active inclusion bodies for d-serine synthesis. Enzym Microb Technol 137:109547
Wang X, Zhou BH, Hu WK et al (2015) Formation of active inclusion bodies induced by hydrophobic self-assembling peptide GFIL8. Microb Cell Fact 14:88
Du J, Rehm BHA (2017) Purification of target proteins from intracellular inclusions mediated by intein cleavable polyhydroxyalkanoate synthase fusions. Microb Cell Factories 16:184
Xing L, Wu W, Zhou B et al (2011) Streamlined protein expression and purification using cleavable self-aggregating tags. Microb Cell Factories 10:42
Villaverde A, Garcia-Fruitos E, Rinas U et al (2012) Packaging protein drugs as bacterial inclusion bodies for therapeutic applications. Microb Cell Factories 11:76
García-Fruitós E, Rodríguez-Carmona E, Díez-Gil C et al (2009) Surface cell growth engineering assisted by a novel bacterial nanomaterial. Adv Mat 21:4249–4253
García-Fruitós E, Seras-Franzoso J, Vazquez E et al (2010) Tunable geometry of bacterial inclusion bodies as substrate materials for tissue engineering. Nanotechnology 21:205101
Gil-Garcia M, Navarro S, Ventura S (2020) Coiled-coil inspired functional inclusion bodies. Microb Cell Factories 19:117
Choi SL, Lee SJ, Ha JS et al (2011) Generation of catalytic protein particles in Escherichia coli cells using the cellulose-binding domain from Cellulomonas fimi as a fusion partner. Biotechnol Bioproc E 16:1173–1179
Nahalka J, Nidetzky B (2007) Fusion to a pull-down domain: a novel approach of producing Trigonopsis variabilis D-amino acid oxidase as insoluble enzyme aggregates. Biotechnol Bioeng 97:454–461
Park SY, Park SH, Choi SK (2012) Active inclusion body formation using Paenibacillus polymyxa PoxB as a fusion partner in Escherichia coli. Anal Biochem 426:63–65
Lin ZL, Zhou BH, Wu W et al (2013) Self-assembling amphipathic alpha-helical peptides induce the formation of active protein aggregates in vivo. Faraday Discuss 166:243–256
Zhou BH, Xing L, Wu W et al (2012) Small surfactant-like peptides can drive soluble proteins into active aggregates. Microb Cell Factories 11:10
Gifre-Renom L, Cano-Garrido O, Fabregas F et al (2018) A new approach to obtain pure and active proteins from Lactococcus lactis protein aggregates. Sci Rep 8:13917
Lamm R, Jager VD, Heyman B et al (2020) Detailed small-scale characterization and scale-up of active YFP inclusion body production with Escherichia coli induced by a tetrameric coiled coil domain. J Biosci Bioeng 129:730–740
Berman HM, Westbrook J, Feng Z et al (2000) The protein data bank. Nucleic Acids Res 28:235–242
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797
Kuriata A, Iglesias V, Pujols J et al (2019) Aggrescan3D (A3D) 2.0: prediction and engineering of protein solubility. Nucleic Acids Res 47:W300–W307
Ferruz N, Schmidt S, Hocker B (2021) ProteinTools: a toolkit to analyze protein structures. Nucleic Acids Res 49:W559–W566
Jacak R, Leaver-Fay A, Kuhlman B (2012) Computational protein design with explicit consideration of surface hydrophobic patches. Proteins 80:825–838
Kuhlman B, Baker D (2000) Native protein sequences are close to optimal for their structures. Proc Natl Acad Sci U S A 97:10383–10388
Rohl CA, Strauss CE, Misura KM et al (2004) Protein structure prediction using Rosetta. Methods Enzymol 383:66–93
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF chimera--a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Schrödinger L, Delano W (2020) PyMOL [Internet]. Available from: http://www.pymol.org/pymol
Studier FW (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234
Fox BG, Blommel PG (2009) Autoinduction of protein expression. Curr Protoc Protein Sci Chapter 5:Unit 5.23
Blommel PG, Becker KJ, Duvnjak P et al (2007) Enhanced bacterial protein expression during auto-induction obtained by alteration of lac repressor dosage and medium composition. Biotechnol Prog 23:585–598
Swinehart DF (1962) The Beer-Lambert Law. J Chem Educ 39:333
Gasteiger E, Hoogland C, Gattiker A et al (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, Totowa, New Jersey, USA, pp 571–607
Hameduh T, Haddad Y, Adam V et al (2020) Homology modeling in the time of collective and artificial intelligence. Comput Struct Biotechnol J 18:3494–3506
Lu Y, Xiao S, Yuan M et al (2018) Using overlap-extension PCR technique to fusing genes for constructing recombinant plasmids. J Basic Microbiol 58:273–276
Engler C, Kandzia R, Marillonnet S (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One 3:e3647
Kulig J, Frese A, Kroutil W et al (2013) Biochemical characterization of an alcohol dehydrogenase from Ralstonia sp. Biotechnol Bioeng 110:1838–1848
Acknowledgments
The research was financially supported by the CLIB Competence Center Biotechnology (CKB) funded by the European Regional Development Fund ERDF (34.EFRE 0300096 and 34.EFRE 0300097).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Ölçücü, G., Jaeger, KE., Krauss, U. (2023). Design, Production, and Characterization of Catalytically Active Inclusion Bodies. In: Kopp, J., Spadiut, O. (eds) Inclusion Bodies. Methods in Molecular Biology, vol 2617. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2930-7_4
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2930-7_4
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2929-1
Online ISBN: 978-1-0716-2930-7
eBook Packages: Springer Protocols