Abstract
Dynamic contrast-enhanced (DCE) MRI monitors the transit of contrast agents, typically gadolinium chelates, through the intrarenal regions, the renal cortex, the medulla, and the collecting system. In this way, DCE-MRI reveals the renal uptake and excretion of the contrast agent. An optimal DCE-MRI acquisition protocol involves finding a good compromise between whole-kidney coverage (i.e., 3D imaging), spatial and temporal resolution, and contrast resolution. By analyzing the enhancement of the renal tissues as a function of time, one can determine indirect measures of clinically important single-kidney parameters as the renal blood flow, glomerular filtration rate, and intrarenal blood volumes. Gadolinium-containing contrast agents may be nephrotoxic in patients suffering from severe renal dysfunction, but otherwise DCE-MRI is clearly useful for diagnosis of renal functions and for assessing treatment response and posttransplant rejection.
Here we introduce the concept of renal DCE-MRI, describe the existing methods, and provide an overview of preclinical DCE-MRI applications to illustrate the utility of this technique to measure renal perfusion and glomerular filtration rate in animal models.
This publication is based upon work from the COST Action PARENCHIMA, a community-driven network funded by the European Cooperation in Science and Technology (COST) program of the European Union, which aims to improve the reproducibility and standardization of renal MRI biomarkers. This introduction is complemented by two separate publications describing the experimental procedure and data analysis.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- Magnetic resonance imaging (MRI)
- Dynamic contrast-enhanced (DCE)
- Kidney
- Mice
- Rats
- T1 mapping
- Contrast agent
- Perfusion
- Glomerular filtration rate (GFR)
1 Introduction
Glomerular filtration rate (GFR) is a standard measure of kidney function. In past decades, simplified plasma clearance employing radiopharmaceuticals (51Cr-EDTA, 125I-iothalamate, 99mTc-DTPA) was introduced and validated to provide very accurate estimates of total GFR [1,2,3,4]. Later, alternative techniques have been evaluated, using minute doses of nonradioactive contrast agents, including iothalamate and iohexol [5]. Plasma clearance of such compounds has also been applied to measure renal function. Renal blood flow (RBF) is another important measure of the functional status of a kidney. The standard tracer-based technique for quantifying RBF is to measure clearance of substances such as hippurate or of radiopharmaceuticals [6].
Dynamic contrast enhanced (DCE-MRI) is an alternative to these techniques that uses gadolinium chelates as contrast agents (CAs), providing functional information analogous to that obtained from radionuclide studies [7, 8]. These chelates are generally considered glomerular filtration markers, since they are removed from the circulation exclusively by glomerular filtration.
DCE-MRI measures T1-weighted signal intensity changes in tissues over time after bolus administration of a contrast agent. Dynamic data acquisition allows for monitoring of the contrast agent in the renal cortex, the medulla, and the collecting system, providing a renographic representation with underlying information about the renal perfusion, filtration, and vascularization [9,10,11,12]. Postprocessing analyses of these signal-vs-time curves reveal kidney functionality in terms of filtration and perfusion, including both heuristic nonmodel approaches and model-based quantitative methods [13, 14]. However, while DCE-MRI is a promising tool in animal models, its clinical utility is hampered by the risk of developing nephrogenic systemic fibrosis in renal patients with severely reduced glomerular filtration capacity [15]. In this chapter, we will address the basic concept of DCE-MRI with the emphasis on kidney disease models in rodents.
This introduction chapter is complemented by two separate chapters describing the experimental procedure and data analysis, which are part of this book.
This chapter is part of the book Pohlmann A, Niendorf T (eds) (2020) Preclinical MRI of the Kidney—Methods and Protocols. Springer, New York.
2 Measurement Concept
2.1 Basic Concept of DCE
The standard acquisition scheme of a DCE-MRI experiment is based on repeated T1-weighted images before and after the injection of a Gadolinium-based CA. In this way, a dynamic curve of the signal enhancement in the kidney is recorded in a time window wide enough (5–10 min) to see the contrast range from the vascular peak enhancement to the wash-out phase. An injection dose of 0.1 mmol Gd/kg is considered standard [16], and lower doses are occasionally exploited to reduce concomitant T2* effects in vessels.
2.2 Gadolinium-Based Contrast Agents
In DCE-MRI, the gadolinium agents have two main advantages, one attributed to the properties of the gadolinium-ion in response to a magnetic field, and one attributed to the chelate that has a biodistribution in the body which favors measurements of renal clearance. In physical terms, the gadolinium ion is chosen having seven unpaired electrons in its orbitals resulting in strong paramagnetism [17,18,19]. Furthermore, the symmetry of the electron configuration of the trivalent gadolinium ion is a hospitable environment for electron spins, contributing to the rise of the crucial nuclear magnetic resonance parameter called relaxation. For these reasons, gadolinium (Gd3+) is the most common metal ion used in paramagnetic contrast agent and its relationship is given by 1/T1 (Measured) = 1/T1 (Water) + r1 [Gd]. The relaxivity is dependent on the magnetic field and temperature [20,21,22]. Due to its toxicity as free ion, Gd is trapped in a molecular structure defined by the closed/open ring that chelates the metal. These macrocyclic and linear CAs may possess different r1, osmolarity, and molecular size [23,24,25]. In addition to clinical contrast agents, several other Gd complexes have been investigated for providing higher relaxivity or contrast efficiency [26,27,28,29,30,31]. All the contrast agents used for mapping kidney perfusion are small molecular weight extracellular agents and are administered intravenously [32].
2.3 Conversion of Signal into Contrast Agent Concentration
Contrary to nuclear medicine modalities, MRI unfortunately does not measure changes in signal intensity in a straightforward way. This means that measurements of injected contrast agent concentration is difficult, which again severely hinders the possibility to calculate important parameters in absolute units such as ml/min or ml/min/g. Previous studies have taken this constraint into account in three different ways: (1) by assuming that the signal intensity is linearly related to the concentration of contrast agent, (2) by making calibration of water doped with contrast agent and then assume the same signal-vs-concentration can be applied in the living tissue, and (3) by introducing the mathematical complex relationship between signal and concentration. These three approaches are here ordered with increasing difficulties in which they can be performed, meaning that the third method is by far the most ambitious. To be able to convert signal intensities into concentration, we present a step-by-procedure. First, the mathematical equation that relates the signal into concentration relies on both well-known constants, the employed MR imaging technique and the pulse sequence parameters (echo-time, recovery time, flip angle, etc.). Due to the paramagnetic properties of the contrast agent, the magnetic relaxation time (T1) decreases in the presence of the contrast agent. Conversion of acquired signal (enhancement) to changes in T1 needs some mathematical understanding. In a simple setup, DCE-MRI is performed with a T1-weighted-2D-spoiled gradient-echo train (Fig. 1).
First, we consider the spoiled gradient echo sequence (TFE) without prepulses. The longitudinal component of the magnetization is denoted Mz, which relaxes back to its equilibrium M0 due to a characteristic relaxation time (T1). The magnetization Mz is derived by the Bloch equation, given as follows:
where M0 denotes the bulk longitudinal magnetization, and T1 is the longitudinal relaxation time. Integration of Eq. 1 returns the time-dependent Mz(t):
Giving the notation that Mz(t = 0) is Mz(0), we define the constraint:
Resolving Eq. 2 gives the following:
The magnetization just before the n’th pulse (denoted as nTR−) can now be determined:
where E = \( {e}^{-\mathrm{TR}/{T}_1} \), and Mz((n − 1)TR−) represents the magnetization just after a time (n − 1)TR. Eq. 5 can be developed recursively:
Herein, the geometric series is assumed:
The steady-state condition can then be described as follows:
Taking into account that steady-state condition must be fulfilled in presence of a delay TD, employed between the TFE train and the subsequent RF pulse, the following must apply:
Now, let us introduce the preparation pulse (with flip-angle β) applied a time TPREP before the excitation pulse. \( {M}_0^{\mathrm{eq}} \) is taken into account by solving:
where Mz(β+) represents the magnetization just after the β pulse. The magnetization before the n’th α-pulse, denoted as Mz(n,α−), is given as the combination of Eqs. 6 and 10:
Therefore, using a heavily T1-weighted pulse sequence , changes in signal intensity are then almost entirely dependent on changes of T1. In these circumstances, it can be shown that Mzeq is almost linearly dependent on T1 relaxation rate. Note again that the specific magnetic relaxivity of the gadolinium-agent (r) defines the characteristic enhancement property by the equation: [Gd] = (1/T1–1/T1(0))/r, where T1(0) is the bulk relaxation time in the tissue without the presence of contrast agent, and [Gd] is the concentration of contrast (gadolinium) agent. In principle, this means that a precontrast measurement of T1 (T1-mapping) should be performed before injection of the contrast agent. One important factor contributing to erroneous measurements of T1(0) of blood is the inflow effect, because the coherent movement of flowing fluid can alter T1 of the signal arising from spins therein. Placing the central-encoding lines within the low blood flow window within the cardiac cycle can, to some extent, minimize the problems of the inflow effect.
2.4 Imaging Readout
For dynamic acquisitions, two factors must be taken into consideration: the time resolution and the contrast resolution required. For quantitative studies of renal perfusion and GFR , based on the first-pass of the contrast agent, a high temporal resolution is particularly important to accurately sample the vascular phase of the kidney (especially for renal perfusion studies) in order to measure the arterial input function (AIF). The AIF is the signal-time-curve usually observed in the suprarenal abdominal aorta or in a renal artery and is used in different kinetic models in order to compensate for the non-instantaneous bolus injected into the blood. The request for a high temporal resolution explains why some studies have been performed using a single slice acquisition scheme and not as multislice or 3D techniques. However, the required extrapolation of functional data from one slice to the whole kidney is not always valid if the renal disease is irregularly or focally distributed.
Most of the exploited sampling techniques are gradient echo imaging, echo-planar imaging and spiral imaging. Differences in the sequence are represented by the ability to cover the image space, from conventional approaches that acquire a portion of the space every TR to entire space acquisition in every TR step. Among them, other subtechniques exist with little changes in the sampling approach like spoiled gradient echo, segmental echo planar imaging, keyhole imaging or parallel imaging [33].
2.4.1 Gradient-Echo
Gradient echo (also “gradient recalled echo,” GRE)-based sequences are the most commonly used sequences for DCE-MRI studies, where fast GRE sequences can be implemented in different ways, including unbalanced, balanced steady-state, and RF-spoiled sequences [34]. Notably, standard (unbalanced) and balanced gradient echo sequences are sensitive to T2 effects that result in signal decreases upon the accumulation of the contrast agent in the region of interest. Therefore, spoiled GRE sequences are mostly used since they are more sensitive to T1 effects leading to higher contrast enhancements. A drawback of these sequences is the low SNR (signal to noise ratio) that can be compensated by the use of 3D acquisitions.
2.4.2 Echo-Planar Imaging
Echo planar imaging (EPI) allows fast image acquisition in combination with high temporal and spatial resolution, with acquisition of several images per second [35]. On the other hand, EPI sequences are difficult to exploit in abdomen and thorax regions since they are sensitive to susceptibility and motion effects. These artifacts can however be reduced by using segmented EPI sequences with a concomitant loss in temporal resolution.
2.4.3 Keyhole Imaging
Since central lines of the K-space are more sensitive to image contrast in comparison to peripheral lines, keyhole imaging is exploited for accelerating imaging acquisition by acquiring multiple central K-space lines. This high-contrast but low-spatial resolution image can then be combined with a high-resolution image to restore the spatial information. Consequently, temporal resolution can be increased up to three- to fourfold, despite some problems that can be encountered in areas with large physiological movements. Several partial K-space sampling techniques have been developed and exploited in a variety of applications [36].
2.5 T1 Mapping
T1 measurements can be performed along the dynamic acquisition (precontrast and postcontrast) at every repetition time or before and after the bolus administration using T1 mapping techniques. Applying a radiofrequency (RF) pulse, spins will change their magnetization status according to the design of the pulse. The magnetization can be inverted or saturated (nulled), and T1-weighted images are acquired after the inversion or time after saturation pulse. Since the T1 calculation is the time that magnetization takes to restore to its equilibrium (which is considered fully restored after 5 times the T1), the two models of RF pulses are called inversion recovery and saturation recovery. Another way to perturb the magnetization is achieved by manipulating the flip angle of the RF pulse at every sequence acquisition, sampling the T1 relaxation for different angle values.
2.5.1 Saturation Recovery
A 90° saturation pulse effectively nulls the magnetization independently of its state before the saturation pulse. In this way, keeping the repetition time lower than the T1, the system is unable to fully recover to equilibrium and is said to be saturated. Thus, the T1 measurements are performed by varying the TR constantly ranging from 0 to, at least, 2–3 times the longest T1 value expected in the sample. The result is that saturation recovery is a very fast method but with the drawback of a limited dynamic range and limited T1 range.
2.5.2 Inversion Recovery
Inversion recovery consists of a 180° inversion pulse which rotates the longitudinal magnetization to give the classical decay with a 90° pulse. The inversion recovery pulse sequence is repeated continuously, each time applying the same inversion pulse, followed by different waiting times (time of inversion). The principle of this method implies that the repetition time of the sequence must be greater than the longest T1 since the magnetization needs to be fully recovered before applying another inversion pulse. Inversion recovery is a method for T1 map acquisition which produces maps with good accuracy and intensity dynamic range but with the drawback of being time-consuming.
2.5.3 Variable Flip Angle (VFA)
Variable flip angle is used to acquire 3D T1 maps, and the technique uses two or more repeated sequences with different flip angles (ranging from a few degrees to even more than 90°), sampling the T1 relaxation for different angle values. The method is relatively sensitive to B1 inhomogeneities; hence a field map should be acquired. Recently, magnetization transfer effects were identified as an additional source of variability in the obtained VFA data, which can be reduced dramatically by use of tailored composite RF pulses that apply the same power to the bound proton pool for all flip angles [37].
2.6 Renal Handling of Contrast Agents
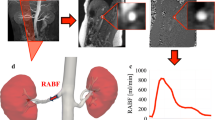
Before analysis of the acquired DCE-MRI data, it is important to understand how the renal segments handle the gadolinium agent during DCE-MRI. Following an i.v. (intravenous) bolus of contrast agent, the arterial concentration is promptly increased following a steady decline over time, reflecting the input and output of the agent over time. Figure 2 shows an example of dynamic enhancement following a bolus of a gadolinium agent, and Fig. 3 shows the handling of a gadolinium agent in the healthy and diseased (ureteric obstructed) kidneys. Graphical presentation of the renal enhancement is denoted as an MR renogram.
Dynamic uptake curve (enhancement) in the aorta (red) and renal whole-parenchyma (yellow) following iv bolus of a gadolinium agent. Purple dots indicate baseline values prior to bolus administration. See Fig. 3 text for MRI sequence parameters. The aorta curve shows a rapid increase followed by a decline over time (0–800 s). The renal curve shows a first-pass vascular uptake, followed by a pattern with decreasing vascular gadolinium-content combined with a glomerular handling of the contrast agent
Example of MR renogram, the result of DCE-MRI of pig kidneys with healthy (left) and diseased (right) kidneys. The yellow ROIs are shown to illustrate where the dynamic curves were drawn. The data were obtained as follows: the anaesthetized pig was placed supine in the magnet, and a surface radiofrequency coil was used for data reception. Fast multislice anatomical images were initially acquired to localize both kidneys. Next, an MRI renography pulse sequence was employed. The whole kidney was covered using a mutlislice (3.0 mm thickness with zero gap) fast 3D gradient echo sequence. Other parameters included: matrix = 128 × 128, field of view = 220 × 220 mm2, TR = 4.3 ms, TE = 1.5 ms. This scan was accompanied by an intravenous injection of 0.05 ml/kg of Gd-DTPA-BMA (Omniscan®; GE Healthcare, Oslo, Norway) was performed as a single bolus administered by hand 10 s after start of a dynamic gradient-echo sequence, with a single phase acquisition time of 2.2 s. A total of 700 dynamic phases were acquired during 7 min
The MR renogram typically consists of vascular, parenchymal, and excretory phases [38]. The vascular phase occurs almost immediately after contrast injection and provides the first segment of the renographic curve, which is reflected by a steep linear rise. In the cortex, this is followed by the parenchymal phase, characterized by continuous uptake, which is represented in the intensity–time curve as a slower linear increase up to a second peak. In the excretory phase, contrast material is released into the collecting system calices and constitutes the third segment of the renographic curve.
2.7 Analysis Methods
While the experimental design of DCE-MRI is simple, extracting true morphological or physiological parameters from a DCE time course is extraordinarily difficult, due to a number of factors, including intravoxel heterogeneity in tissue microstructure and the need for pharmacokinetic (model-based) or empirical (non-model based) modeling of the DCE-MRI signal [16, 38]. Tissue heterogeneity depends upon the organ of interest and image resolution, which are typically not varied in a single experiment. Pharmacokinetic and empirical modeling, however, are active areas of investigation.
2.7.1 Semiquantitative Analysis
As a descriptive approach, the dynamic time-vs-time curve of DCE-MRI data provides information with parameters like time-to-peak enhancement, uptake slope, area-under-the-curve and wash-out rate. These heuristic enhancement parameters may have some relation to physiologic parameters although they also depend on the particular MRI system and acquisition settings, including pulse sequence , sequence parameters, system manufacturer, and contrast agent employed [39]. Figure 4 shows an example of such parameters.
Example of parameters estimated from DCE-MRI in kidneys. Top row from left to right: plasma flow (FP), plasma volume (VP), plasma mean transit time (PMTT). Lower row from left to right: extraction fraction (E), permeable surface area product (PS), and tubular mean transit time (TMMT). The maps are reproduced with permission from Zöllner et al. [39]
2.7.2 Quantitative—Pharmacokinetic Models
Alternatively, various physiological pharmacokinetic models have been proposed, usually based on long-term experiences in nuclear medicine. These models are often attributed to situations where the capillary bed (e.g., the blood–brain barrier) is compromised, resulting in extravasation of the contrast agent through the leaky capillaries. For example, modeling the dynamic extravasation of MRI contrast agent can provide a measure of the extravascular uptake. Overall, applying pharmacokinetic models allow calculation of several measures: (1) physiologic properties (transendothelial permeability, capillary surface area, lesion leakage space), (2) pharmacokinetic parameters (compartmental transfer and rate constants, leakage space), and (3) pathological measures (microvessel density and vascular endothelial growth factor) [13, 16]. Measurements of these parameters using modeling are now considered diagnostically important in tumor evaluation (grade classification) and to recognize the onset of stroke by indirect measurements of rCBF, rCBV, and MTT, although they usually provide limited insight into the underlying pathophysiology of the brain tumor or stroke [40]. It is therefore likely that similar parameters can be measured in the kidney. The quantitative models require additional postprocessing steps, including contrast agent concentration calculation (e.g., based on linearity assumption between concentration and signal changes or in previously acquired T1 maps), selection of a major artery to derive the AIF curve, selection of the pharmacokinetic model (as well as of the number of the compartments and how they are connected) to be used. In addition, the focus of the application as well as the quality of the data will guide the selection of the more appropriate model. Besides, the temporal resolution, protocol injection, and acquisition time can influence the quality of the outcomes, making it necessary to have appropriate and optimized measurement protocols.
2.7.3 Model Selection
All data analysis, including analysis of DCE-MRI data, involves either explicit or implicit comparison to a model. The data are typically considered to be the sum of the signal and noise, and the estimated signal model parameters are of primary interest. In the ideal case, the underlying principles of the measurement and the signal response form the basis of the signal model. Often, however, there are several potential signal models. The analyst must decide which of these competing models best represents the data without “over fitting” (i.e., fitting the noise). Even when the correct signal model is known, the signal-to-noise ratio of the data may not support the complexity of the correct model, requiring that simpler models be considered.
Here, we consider the problem of model selection in DCE-MRI experiments in the kidney. The importance of selecting an appropriate DCE tracer kinetic model to measure tissue perfusion and capillary permeability has already been examined in tumors in previous reports [41, 42]. Model selection algorithms, including Chi-square [43], Akaike information criterion (AIC) [44,45,46], F-test [47,48,49], and the Durbin–Watson statistic [45, 50], have been applied to evaluate tracer kinetic models commonly used in DCE-MRI.
2.7.4 Bayesian Probability Theory
An elegant solution to the model selection problem using Bayesian probability theory-based methods has recently been described [51,52,53]. Bayesian probability theory [54] provides a rigorous formalism for model selection. To determine the optimal model amongst a cohort of competing models, it is necessary to balance the accuracy with which the model recapitulates the data (the goodness of fit, characterized by the residuals) against the number of free parameters in each model (the complexity). Cox’s theorem [55] and its further elaboration by Jaynes [56], state that Bayesian probability theory is the only method of ranking hypotheses concerning model selection that is consistent and can incorporate all of the available prior information. Advances in computational power and the development of Markov-chain Monte Carlo (MCMC) methods have greatly increased the applicability of Bayesian inference to a range of problems.
Bayesian probability theory seeks to assign a probability to the truth of a specific hypothesis. Using a Bayesian approach, the model selection problem treats the model itself as a parameter, for which the posterior probability of a model, given the data and prior information, is computed [51,52,53]. The posterior probability of each model is calculated by:
where P(M|DI) is the posterior probability for any one of M models. The vertical bar “|” means “given the data, D and the prior information, I.” On the right-hand side, P(M|I) is the prior probability for each of the M different models, assigned in this calculation as a uniform prior probability. P(D|MI) is the probability for the data, given model M and prior information I. Finally, the denominator, P(D|I) is a normalization constant that ensures the total probability over all of the models sums to one.
An example of a joint Bayesian approach to model selection in DCE-MRI was described recently by Beeman et al. [57], applied to models of blood flow in cortex in a mouse model of three different renal perfusion rates. The three cohorts of mice studied were: (1) Control; (2) Losartan-treated (high renal blood flow); (3) L-NAME-treated (low renal blood flow). DCE-MRI data were analyzed on a voxel-by-voxel basis. Four models and model parameters were compared in each voxel. Two of the models, the Patlak–Rutland model [58] and the two-compartment model were pharmacokinetic and two, the cumulative log-logistic model with either monoexponential decay with a constant offset or biexponential decay, were empirical. A Markov-chain Monte Carlo simulation was run to compute the posterior probability for the parameters and the model. The estimated probabilities of the parameters of the biexponential model, obtained using this approach, are shown in Fig. 5.
(a, c) Bayesian-estimated posterior probability densities of the empirical biexponential model’s joint fractional amplitudes of the washout and joint slow decay-rate constants, respectively, for each cohort. The Losartan-treated (high renal blood flow) group is represented by the solid black line, the L-NAME (low renal blood flow group) by the dashed black line, and the control group by the dotted gray line. (b, d) The difference in the posterior probability distributions for the joint fractional amplitudes of the washout and joint slow decay-rate constants, respectively, calculated on the high and low flow cohorts. From among the empirical/biexponential model joint parameter estimates, the fractional amplitudes of the washout terms (a) and the slow decay-rate constants (c) differed between mouse cohorts of high and low flow; that is, the 95% confidence interval of the difference in the probability distributions did not overlap with 0 (b and d). (Adapted, with permission, from Beeman, et al. [57])
The importance of signal-to-noise in model selection in DCE-MRI was described recently by Duan et al. [51] In their paper, the authors considered four established kinetic models for analyzing DCE-MRI data collected from patients enrolled in the EMBRACE study, an international study of MRI-guided brachytherapy in locally advanced cervical cancer. The models were (1) Toft’s (TM), (2) Extended Toft’s (ETM), (3) Two-Site Exchange (2CXM), and (4) Compartment Tissue Uptake (CTUM). To demonstrate the importance of signal-to-noise in DCE data analysis, noise-free in silico DCE data sets were generated for each of the four models. The temporal resolution and total data acquisition times were identical to those of the clinical DCE-MRI protocol, using patient-derived tissue parameters.
As expected, Bayesian model selection chose the “correct” signal model for each of these noise-free data sets. Increasing amounts of normally distributed (Gaussian) noise were then added to each data set. At each noise power, 100 independent MCMC simulations (i.e., different noise sets) were performed and the number of “correct” selections, in which model selection chose the model used to simulate the data, was recorded. The number of correct model selections varied as a function of both model complexity and noise power. With increasing levels of noise, simpler data representations were preferred relative to more complex models (Fig. 6a). Figure 6b shows model selection results for in silico data created based on the 2XCM. The number of correct model selections dropped rapidly with increasing noise, with a concomitant increase in the selection of the CTUM, a simplified version of the 2XCM. Figure 6c, d shows how the accuracy and uncertainty of estimations of extravascular, extracellular volume (ve), the only common parameter between the simplest model (TM) and the most complex model (2XCM), vary with added noise. At every noise level, the uncertainties of the estimated ve are much smaller for the TM, relative to the 2CXM.
Bayesian DCE-MRI model-selection and parameter-estimation results for in silico datasets created based on the whole-tumor averaged ROI DCE time course from a single patient in the EMBRACE study. (a) For each of the four in silico DCE-MRI data models, the number of “correct” model selections (out of 100 different noise representations), as a function of the noise standard deviation (SD). (b) For the two-compartment exchange in silico DCE-MRI data model (the most complex model of the four models examined), the number of times a given model was selected (out of 100 different noise representations) as a function of the noise SD. (c, d) Relative percent error of ve (extracellular-extravascular volume fraction) estimated from initially noiseless simulated TM (c) and 2CXM (d) data as a function of added noise. (Adapted with permission from Duan et al. [51])
2.8 Practical Considerations
The main advantage of T1-weighted DCE-MRI is that both tissue blood flow and filtration (permeability surface) can be measured simultaneously. One important element in the T1-weighted DCE-MRI is the underlying model from which the calculated parameters are derived. Many assumptions about the tissue system are made with respect to the nature of contrast agent kinetic modeling, and these assumptions may affect the accuracy of the parameter calculations. In practice, a compromise must be struck between reality and the precision of the measured data (signal-to noise ratio, temporal resolution, etc.). The different models proposed by leading experts in this field have unfortunately been presented with a variety of quantities, meaning that comparisons between different groups are almost impossible. Consequently, a standardization of quantities associated with analysis of T1-weighted DCE-MRI has been proposed in the review by Tofts et al. [59], and later by Sourbron [42] as part of building up a common language for the estimation and description of physiologic parameters in the kidney. For all methods proposed, the calculation of renal parameters requires knowledge of the arterial input function, which in practice is derived from the abdominal aorta or a renal artery with the assumption that this represents the exact input to the renal tissue. Delay and dispersion of the bolus that are introduced during its passage from the site of arterial input function estimation to the renal tissue will therefore introduce an error in the quantification of RBF or GFR , and this error could very well vary from one region to another because of the differences in the amount of delay and dispersion for different kidney regions. Thus, some general approximations seem useful: (1) DCE-MRI is performed under a steady-state condition. (2) The renal relaxivity of the gadolinium-agent is known. (3) Some conversion to gadolinium concentration can be made. (4) An input curve is defined. (5) A mathematical analytical model can be approximated.
Today, little is known about the potential of gadolinium-enhanced MRI for the assessment of the regional renal blood flow and the regional glomerular filtration rate. Nor do we know the limitations and accuracy of this technique under different pathological and pathophysiological conditions. Thirdly, we do not know the clinical applicability of DCE-MRI and the role it may have in clinical diagnosis. A step-by-step procedure should therefore be initiated to exploit the abovementioned fundamental questions: (1) The methodologies should be further validated against standard methods to obtain reproducible results in agreements to those obtained by clinical non-MRI techniques. (2) Further animal studies should be performed to examine the accuracy and availability under different pathological and pathophysiological conditions. (3) Experimental studies in humans, where the DCE-MRI methods are validated against a standard reference method.
3 Overview of Applications
3.1 DCE-MRI Validation Studies in Animal Models
Several studies investigated DCE-MRI approaches for assessing renal filtration in animal models. Annet et al. estimated the GFR in rabbit kidneys by applying a compartment model and compared the MRI-derived estimate with the measured plasma clearance of 51Cr-EDTA [60]. Although MRI-based GFR values were lower than those measured experimentally, a marked correlation was observed between the two techniques. Winter et al. compared renal perfusion values obtained by arterial spin labeling (ASL) or DCE-MRI in healthy rats, with absolute renal cortex perfusion estimates in close agreement with those obtained by ASL [61].
Zimmer et al. performed an inter- and intramethodical comparison by measuring differences in renal blood flow (RBF) in five rats with unilateral ischemic acute kidney injury with both ASL and DCE-MRI. Both, the FAIR-ASL approach and DCE-MRI deconvolution technique showed significant differences in RBF between healthy and diseased kidneys as shown in Fig. 7 [62]. The validation of MRI-based GFR estimation by optical imaging was proposed by Sadick et al. [63]. GFR estimation was performed by DCE-MRI on a 3 T whole body scanner using a dedicated animal rat volume coil and by clearance using a fluorescent tracer (FITC-sinistrin) and an optical imaging device. However, the correlation between optical GFR and MRI-based GFR was poor, probably due to the fact that measurements were performed on different days involving two anesthesia thus affecting the physiology of the animals. Zöllner et al. recently showed a simultaneous measurement of optical and MR-based GFR in healthy rats and rats with unilateral nephrectomy (UNX) [64]. A two-compartment filtration model was employed to calculate a map of the tubular flow. Subsequent, taking the cortex volume into account single kidney GFR values were calculated with a good correlation observed between the two methods. The reduction of GFR in the UNX rats was about 50% and observed for both techniques.
Exemplary illustration of perfusion MRI of a rat with left-side AKI and perfusion maps. (a) True-FISP M0 image of an ASL measurement and the corresponding perfusion map (b). (c) TWIST post–contrast agent injection image and the corresponding RBF map (d). All drawings show the same rat and the same axial slice. Differences between the kidney with AKI and the contralateral kidney are clearly visible on the MRI images as well as on the perfusion maps. (Reproduced with permission from Zimmer et al. [62])
The role played by the binding to serum proteins in the calculation of DCE-MRI derived estimates was investigated by Notohamiprodjo et al. by administering Gadolinium-based contrast agents with different chemical structure [65]. The two investigated contrast agents, Gd-BOPTA and Gd-DTPA differ in their binding affinity toward serum albumin, hence affecting both the relaxivity and the pharmacokinetic properties [66,67,68]. In fact, binding to the serum protein results in longer circulation times (hence reduced filtration) and higher contrast capability that are easily observed at low magnetic fields (0.5–1.5 T) [69,70,71,72,73]. Therefore, whereas similar renal perfusion values were obtained by both the two contrast agents, GFR estimates were not accurate when obtained with the albumin-binding one (Gd-BOPTA), thus indicating that even low albumin binding may affect GFR estimates.
3.2 DCE-MRI for Assessing Kidney Damages in Animal Models
In the preclinical panorama, DCE-MRI has been widely used to assess kidney function in renovascular and parenchymal diseases that may lead to chronic kidney failure but also in acute events like ischemia–reperfusion, toxicity, infections, or surgery. Thus, different animal models have been explored ranging from classical acute kidney injury (AKI) and chronic kidney disease (CKD) to posttransplant rejection, nephrectomy, and assessment of drug treatment [74,75,76,77,78,79,80]. In a murine model of unilateral renal artery stenosis (RAS), measurements of single kidney GFR and perfusion by a modified two-compartment model were compared to those obtained from fluorescein isothiocyanate (FITC)-inulin clearance and ASL , respectively. Both renal GFR and perfusion measurements were in close agreement with the FITC-inulin clearance and ASL methods, with a marked reduction in GFR and perfusion estimates for the stenotic as compared to the control kidneys [81]. Ischemia–reperfusion injury has been exploited to model acute kidney damages and DCE-MRI approaches have been investigated for assessing single-kidney function in both mice and rats [82, 83]. Significant reduction in single kidney GFR (calculated by applying a two compartment renal filtration model) was observed for ischemic kidneys in comparison to contralateral ones in rats [84]. Conversely, for all the other DCE-MRI derived functional parameters (mean transit times and renal blood volume) differences were present, but were not significant between diseased and contralateral kidneys of ischemic rats.
DCE-MRI studies have also been exploited in murine renal transplantations studies for assessing acute rejection and evaluation of immunosuppressive therapy [85]. In this study, a deconvolution approach was applied to the DCE-MRI data and the plasma flow estimate provided restored organ perfusion under Ciclosporin A therapy in syngeneic transplants compared to controls.
References
Hauser W, Atkins HL, Nelson KG, Richards P (1970) Technetium-99m DTPA: a new radiopharmaceutical for brain and kidney scanning. Radiology 94(3):679–684. https://doi.org/10.1148/94.3.679
Taylor A (1999) Radionuclide renography: a personal approach. Semin Nucl Med 29(2):102–127
Gaspari F, Perico N, Remuzzi G (1997) Measurement of glomerular filtration rate. Kidney Int Suppl 63:S151–S154
Barbour GL, Crumb CK, Boyd CM, Reeves RD, Rastogi SP, Patterson RM (1976) Comparison of inulin, iothalamate, and 99mTc-DTPA for measurement of glomerular filtration rate. J Nucl Med 17(4):317–320
Soveri I, Berg UB, Bjork J, Elinder CG, Grubb A, Mejare I, Sterner G, Back SE, SBU GFR Review Group (2014) Measuring GFR: a systematic review. Am J Kidney Dis 64(3):411–424. https://doi.org/10.1053/j.ajkd.2014.04.010
Peters AM (1991) Quantification of renal haemodynamics with radionuclides. Eur J Nucl Med 18(4):274–286
Suto Y, Caner BE, Tamagawa Y, Matsuda T, Nakashima T, Matsushita T, Odori T, Ishii Y, Torizuka K (1989) Assessment of magnetic resonance contrast enhancement with Gd-DTPA: comparison with the uptake of Tc-99m-DTPA. Radiat Med 7(5):209–213
Prato FS, Wisenberg G, Marshall TP, Uksik P, Zabel P (1988) Comparison of the biodistribution of gadolinium-153 DTPA and technetium-99m DTPA in rats. J Nucl Med 29(10):1683–1687
Choyke PL, Frank JA, Girton ME, Inscoe SW, Carvlin MJ, Black JL, Austin HA, Dwyer AJ (1989) Dynamic Gd-DTPA-enhanced MR imaging of the kidney: experimental results. Radiology 170(3 Pt 1):713–720. https://doi.org/10.1148/radiology.170.3.2916025
Fransen R, Muller HJ, Boer WH, Nicolay K, Koomans HA (1996) Contrast-enhanced dynamic magnetic resonance imaging of the rat kidney. J Am Soc Nephrol 7(3):424–430
Wen JG, Chen Y, Ringgaard S, Frokiaer J, Jorgensen TM, Stodkilde-Jorgensen H, Djurhuus JC (2000) Evaluation of renal function in normal and hydronephrotic kidneys in rats using gadolinium diethylenetetramine-pentaacetic acid enhanced dynamic magnetic resonance imaging. J Urol 163(4):1264–1270
Kikinis R, von Schulthess GK, Jager P, Durr R, Bino M, Kuoni W, Kubler O (1987) Normal and hydronephrotic kidney: evaluation of renal function with contrast-enhanced MR imaging. Radiology 165(3):837–842. https://doi.org/10.1148/radiology.165.3.3685363
Sourbron SP, Michaely HJ, Reiser MF, Schoenberg SO (2008) MRI-measurement of perfusion and glomerular filtration in the human kidney with a separable compartment model. Investig Radiol 43(1):40–48. https://doi.org/10.1097/RLI.0b013e31815597c5
Pedersen M, Shi Y, Anderson P, Stodkilde-Jorgensen H, Djurhuus JC, Gordon I, Frokiaer J (2004) Quantitation of differential renal blood flow and renal function using dynamic contrast-enhanced MRI in rats. Magn Reson Med 51(3):510–517. https://doi.org/10.1002/mrm.10711
Elmholdt TR, Pedersen M, Jorgensen B, Sondergaard K, Jensen JD, Ramsing M, Olesen AB (2011) Nephrogenic systemic fibrosis is found only among gadolinium-exposed patients with renal insufficiency: a case-control study from Denmark. Br J Dermatol 165(4):828–836. https://doi.org/10.1111/j.1365-2133.2011.10465.x
Grenier N, Mendichovszky I, de Senneville BD, Roujol S, Desbarats P, Pedersen M, Wells K, Frokiaer J, Gordon I (2008) Measurement of glomerular filtration rate with magnetic resonance imaging: principles, limitations, and expectations. Semin Nucl Med 38(1):47–55. https://doi.org/10.1053/j.semnuclmed.2007.09.004
Aime S, Botta M, Terreno E (2005) Gd(III)-based contrast agents for MRI. Adv Inorg Chem 57(57):173–237. https://doi.org/10.1016/S0898-8838(05)57004-1
Caravan P, Ellison JJ, McMurry TJ, Lauffer RB (1999) Gadolinium(III) chelates as MRI contrast agents: structure, dynamics, and applications. Chem Rev 99(9):2293–2352
Aime S, Botta M, Fasano M, Crich SG, Terreno E (1996) Gd(III) complexes as contrast agents for magnetic resonance imaging: a proton relaxation enhancement study of the interaction with human serum albumin. J Biol Inorg Chem 1(4):312–319. https://doi.org/10.1007/s007750050059
Lauffer RB (1987) Paramagnetic metal complexes as water proton relaxation agents for NMR imaging: theory and design. Chem Rev 87(5):901–927
Werner EJ, Datta A, Jocher CJ, Raymond KN (2008) High-relaxivity MRI contrast agents: where coordination chemistry meets medical imaging. Angew Chem Int Ed Engl 47(45):8568–8580. https://doi.org/10.1002/anie.200800212
Burtea C, Laurent S, Vander Elst L, Muller RN (2008) Contrast agents: magnetic resonance. Handb Exp Pharmacol (185 Pt 1):135-165. https://doi.org/10.1007/978-3-540-72718-7_7
Aime S, Caravan P (2009) Biodistribution of gadolinium-based contrast agents, including gadolinium deposition. J Magn Reson Imaging 30(6):1259–1267. https://doi.org/10.1002/jmri.21969
Baranyai Z, Palinkas Z, Uggeri F, Maiocchi A, Aime S, Brucher E (2012) Dissociation kinetics of open-chain and macrocyclic gadolinium(III)-aminopolycarboxylate complexes related to magnetic resonance imaging: catalytic effect of endogenous ligands. Chemistry 18(51):16426–16435. https://doi.org/10.1002/chem.201202930
Baranyai Z, Uggeri F, Giovenzana GB, Benyei A, Brucher E, Aime S (2009) Equilibrium and kinetic properties of the lanthanoids(III) and various divalent metal complexes of the heptadentate ligand AAZTA. Chemistry 15(7):1696–1705. https://doi.org/10.1002/chem.200801803
Sherry AD, Caravan P, Lenkinski RE (2009) Primer on gadolinium chemistry. J Magn Reson Imaging 30(6):1240–1248. https://doi.org/10.1002/jmri.21966
Wahsner J, Gale EM, Rodriguez-Rodriguez A, Caravan P (2019) Chemistry of MRI contrast agents: current challenges and new frontiers. Chem Rev 119(2):957–1057. https://doi.org/10.1021/acs.chemrev.8b00363
Dastru W, Longo D, Aime S (2011) Contrast agents and mechanisms. Drug Discov Today 8(2–4):e109–e115. https://doi.org/10.1016/j.ddtec.2011.11.013
Terreno E, Dastru W, Delli Castelli D, Gianolio E, Geninatti Crich S, Longo D, Aime S (2010) Advances in metal-based probes for MR molecular imaging applications. Curr Med Chem 17(31):3684–3700
Avedano S, Botta M, Haigh JS, Longo DL, Woods M (2013) Coupling fast water exchange to slow molecular tumbling in Gd3+ chelates: why faster is not always better. Inorg Chem 52(15):8436–8450. https://doi.org/10.1021/ic400308a
Longo DL, Arena F, Consolino L, Minazzi P, Geninatti-Crich S, Giovenzana GB, Aime S (2016) Gd-AAZTA-MADEC, an improved blood pool agent for DCE-MRI studies on mice on 1 T scanners. Biomaterials 75:47–57. https://doi.org/10.1016/j.biomaterials.2015.10.012
Pierre VC, Allen MJ, Caravan P (2014) Contrast agents for MRI: 30+ years and where are we going? J Biol Inorg Chem 19(2):127–131. https://doi.org/10.1007/s00775-013-1074-5
Paschal CB, Morris HD (2004) K-space in the clinic. J Magn Reson Imaging 19(2):145–159. https://doi.org/10.1002/jmri.10451
Markl M, Leupold J (2012) Gradient echo imaging. J Magn Reson Imaging 35(6):1274–1289. https://doi.org/10.1002/jmri.23638
Tsao J (2010) Ultrafast imaging: principles, pitfalls, solutions, and applications. J Magn Reson Imaging 32(2):252–266. https://doi.org/10.1002/jmri.22239
Oesterle C, Strohschein R, Kohler M, Schnell M, Hennig J (2000) Benefits and pitfalls of keyhole imaging, especially in first-pass perfusion studies. J Magn Reson Imaging 11(3):312–323
Teixeira RPAG, Malik SJ, Hajnal JV (2019) Fast quantitative MRI using controlled saturation magnetization transfer. Magn Reson Med 81(2):907–920. https://doi.org/10.1002/mrm.27442
Grenier N, Pedersen M, Hauger O (2006) Contrast agents for functional and cellular MRI of the kidney. Eur J Radiol 60(3):341–352. https://doi.org/10.1016/j.ejrad.2006.06.024
Zollner FG, Daab M, Sourbron SP, Schad LR, Schoenberg SO, Weisser G (2016) An open source software for analysis of dynamic contrast enhanced magnetic resonance images: UMMPerfusion revisited. BMC Med Imaging 16:7. https://doi.org/10.1186/s12880-016-0109-0
Roldan-Valadez E, Gonzalez-Gutierrez O, Martinez-Lopez M (2012) Diagnostic performance of PWI/DWI MRI parameters in discriminating hyperacute versus acute ischaemic stroke: finding the best thresholds. Clin Radiol 67(3):250–257. https://doi.org/10.1016/j.crad.2011.08.020
Ewing JR, Bagher-Ebadian H (2013) Model selection in measures of vascular parameters using dynamic contrast-enhanced MRI: experimental and clinical applications. NMR Biomed 26(8):1028–1041. https://doi.org/10.1002/nbm.2996
Sourbron SP, Buckley DL (2013) Classic models for dynamic contrast-enhanced MRI. NMR Biomed 26(8):1004–1027. https://doi.org/10.1002/nbm.2940
Abrikossova N, Skoglund C, Ahren M, Bengtsson T, Uvdal K (2012) Effects of gadolinium oxide nanoparticles on the oxidative burst from human neutrophil granulocytes. Nanotechnology 23(27):275101. https://doi.org/10.1088/0957-4484/23/27/275101
Kallehauge JF, Tanderup K, Duan C, Haack S, Pedersen EM, Lindegaard JC, Fokdal LU, Mohamed SM, Nielsen T (2014) Tracer kinetic model selection for dynamic contrast-enhanced magnetic resonance imaging of locally advanced cervical cancer. Acta Oncol 53(8):1064–1072. https://doi.org/10.3109/0284186X.2014.937879
Li X, Welch EB, Chakravarthy AB, Xu L, Arlinghaus LR, Farley J, Mayer IA, Kelley MC, Meszoely IM, Means-Powell J, Abramson VG, Grau AM, Gore JC, Yankeelov TE (2012) Statistical comparison of dynamic contrast-enhanced MRI pharmacokinetic models in human breast cancer. Magn Reson Med 68(1):261–271. https://doi.org/10.1002/mrm.23205
Naish JH, Kershaw LE, Buckley DL, Jackson A, Waterton JC, Parker GJ (2009) Modeling of contrast agent kinetics in the lung using T1-weighted dynamic contrast-enhanced MRI. Magn Reson Med 61(6):1507–1514. https://doi.org/10.1002/mrm.21814
Bagher-Ebadian H, Jain R, Nejad-Davarani SP, Mikkelsen T, Lu M, Jiang Q, Scarpace L, Arbab AS, Narang J, Soltanian-Zadeh H, Paudyal R, Ewing JR (2012) Model selection for DCE-T1 studies in glioblastoma. Magn Reson Med 68(1):241–251. https://doi.org/10.1002/mrm.23211
Chwang WB, Jain R, Bagher-Ebadian H, Nejad-Davarani SP, Iskander AS, VanSlooten A, Schultz L, Arbab AS, Ewing JR (2014) Measurement of rat brain tumor kinetics using an intravascular MR contrast agent and DCE-MRI nested model selection. J Magn Reson Imaging 40(5):1223–1229. https://doi.org/10.1002/jmri.24469
Donaldson SB, West CM, Davidson SE, Carrington BM, Hutchison G, Jones AP, Sourbron SP, Buckley DL (2010) A comparison of tracer kinetic models for T1-weighted dynamic contrast-enhanced MRI: application in carcinoma of the cervix. Magn Reson Med 63(3):691–700. https://doi.org/10.1002/mrm.22217
Draper NR, Smith H (1998) Applied regression analysis, 3rd edn. Wiley, New York
Duan C, Kallehauge JF, Bretthorst GL, Tanderup K, Ackerman JJ, Garbow JR (2017) Are complex DCE-MRI models supported by clinical data? Magn Reson Med 77(3):1329–1339. https://doi.org/10.1002/mrm.26189
Meinerz K, Beeman SC, Duan C, Bretthorst GL, Garbow JR, Ackerman JJH (2018) Bayesian modeling of NMR data: quantifying longitudinal relaxation in vivo, and in vitro with a tissue-water-relaxation mimic (crosslinked bovine serum albumin). Appl Magn Reson 49(1):3–24
Quirk JD, Bretthorst GL, Garbow JR, Ackerman JJ (2019) Magnetic resonance data modeling: the Bayesian analysis toolbox. Concepts Magn Reson Part A 47A:e21467
Bayes T, Price R (1763) An essay toward solving a problem in the doctrine of chance. Philos Trans R Soc Lond 53:370–418
Cox RT (1961) The algebra of probable inference. Johns Hopkins University Press, Baltimore, MD
Jaynes ET (1988) Optimal information-processing and Bayes theorem - comment. Am Stat 42(4):280–281. https://doi.org/10.2307/2685144
Beeman SC, Osei-Owusu P, Duan C, Engelbach J, Bretthorst GL, Ackerman JJH, Blumer KJ, Garbow JR (2015) Renal DCE-MRI model selection using Bayesian probability theory. Tomography 1(1):61–68. https://doi.org/10.18383/j.tom.2015.00133
Patlak CS, Blasberg RG, Fenstermacher JD (1983) Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J Cereb Blood Flow Metab 3(1):1–7. https://doi.org/10.1038/jcbfm.1983.1
Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp MV, Larsson HB, Lee TY, Mayr NA, Parker GJ, Port RE, Taylor J, Weisskoff RM (1999) Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging 10(3):223–232. https://doi.org/10.1002/(SICI)1522-2586(199909)10:3<223::AID-JMRI2>3.0.CO;2-S
Annet L, Hermoye L, Peeters F, Jamar F, Dehoux JP, Van Beers BE (2004) Glomerular filtration rate: assessment with dynamic contrast-enhanced MRI and a cortical-compartment model in the rabbit kidney. J Magn Reson Imaging 20(5):843–849. https://doi.org/10.1002/jmri.20173
Winter JD, St Lawrence KS, Cheng HL (2011) Quantification of renal perfusion: comparison of arterial spin labeling and dynamic contrast-enhanced MRI. J Magn Reson Imaging 34(3):608–615. https://doi.org/10.1002/jmri.22660
Zimmer F, Zollner FG, Hoeger S, Klotz S, Tsagogiorgas C, Kramer BK, Schad LR (2013) Quantitative renal perfusion measurements in a rat model of acute kidney injury at 3T: testing inter- and intramethodical significance of ASL and DCE-MRI. PLoS One 8(1):e53849. https://doi.org/10.1371/journal.pone.0053849
Sadick M, Attenberger U, Kraenzlin B, Kayed H, Schoenberg SO, Gretz N, Schock-Kusch D (2011) Two non-invasive GFR-estimation methods in rat models of polycystic kidney disease: 3.0 tesla dynamic contrast-enhanced MRI and optical imaging. Nephrol Dial Transplant 26(10):3101–3108. https://doi.org/10.1093/ndt/gfr148
Zollner FG, Schock-Kusch D, Backer S, Neudecker S, Gretz N, Schad LR (2013) Simultaneous measurement of kidney function by dynamic contrast enhanced MRI and FITC-sinistrin clearance in rats at 3 tesla: initial results. PLoS One 8(11):e79992. https://doi.org/10.1371/journal.pone.0079992
Notohamiprodjo M, Pedersen M, Glaser C, Helck AD, Lodemann K-P, Jespersen B, Fischereder M, Reiser MF, Sourbron SP (2011) Comparison of Gd-DTPA and Gd-BOPTA for studying renal perfusion and filtration. J Magn Reson Imaging 34(3):595–607. https://doi.org/10.1002/jmri.22640
Laurent S, Elst LV, Muller RN (2006) Comparative study of the physicochemical properties of six clinical low molecular weight gadolinium contrast agents. Contrast Media Mol Imaging 1(3):128–137. https://doi.org/10.1002/cmmi.100
Caravan P, Cloutier NJ, Greenfield MT, McDermid SA, Dunham SU, Bulte JW, Amedio JC Jr, Looby RJ, Supkowski RM, Horrocks WD Jr, McMurry TJ, Lauffer RB (2002) The interaction of MS-325 with human serum albumin and its effect on proton relaxation rates. J Am Chem Soc 124(12):3152–3162
Aime S, Barge A, Botta M, Terreno E (2003) Interactions of lanthanides and their complexes with proteins. Conclusions regarding magnetic resonance imaging. Met Ions Biol Syst 40:643–682
Caravan P, Parigi G, Chasse JM, Cloutier NJ, Ellison JJ, Lauffer RB, Luchinat C, McDermid SA, Spiller M, McMurry TJ (2007) Albumin binding, relaxivity, and water exchange kinetics of the diastereoisomers of MS-325, a gadolinium(III)-based magnetic resonance angiography contrast agent. Inorg Chem 46(16):6632–6639. https://doi.org/10.1021/ic700686k
Botta M, Avedano S, Giovenzana GB, Lombardi A, Longo D, Cassino C, Tei L, Aime S (2011) Relaxometric study of a series of monoaqua Gd-III complexes of rigidified EGTA-like chelators and their noncovalent interaction with human serum albumin. Eur J Inorg Chem (6):802–810. https://doi.org/10.1002/ejic.201001103
Gianolio E, Giovenzana GB, Longo D, Longo I, Menegotto I, Aime S (2007) Relaxometric and modelling studies of the binding of a lipophilic Gd-AAZTA complex to fatted and defatted human serum albumin. Chemistry 13(20):5785–5797. https://doi.org/10.1002/chem.200601277
Avedano S, Tei L, Lombardi A, Giovenzana GB, Aime S, Longo D, Botta M (2007) Maximizing the relaxivity of HSA-bound gadolinium complexes by simultaneous optimization of rotation and water exchange. Chem Commun (Camb) (45):4726–4728. https://doi.org/10.1039/b714438e
Aime S, Gianolio E, Longo D, Pagliarin R, Lovazzano C, Sisti M (2005) New insights for pursuing high relaxivity MRI agents from modelling the binding interaction of Gd(III) chelates to HSA. Chembiochem 6(5):818–820. https://doi.org/10.1002/cbic.200400364
Egger C, Cannet C, Gérard C, Debon C, Stohler N, Dunbar A, Tigani B, Li J, Beckmann N (2015) Adriamycin-induced nephropathy in rats: functional and cellular effects characterized by MRI. J Magn Reson Imaging 41(3):829–840. https://doi.org/10.1002/jmri.24603
Laurent D, Poirier K, Wasvary J, Rudin M (2002) Effect of essential hypertension on kidney function as measured in rat by dynamic MRI. Magn Reson Med 47(1):127–134
Hermoye L, Annet L, Lemmerling P, Peeters F, Jamar F, Gianello P, Van Huffel S, Van Beers BE (2004) Calculation of the renal perfusion and glomerular filtration rate from the renal impulse response obtained with MRI. Magn Reson Med 51(5):1017–1025. https://doi.org/10.1002/mrm.20026
Zhang YD, Wang J, Zhang J, Wang X, Jiang X (2014) Effect of iodinated contrast media on renal function evaluated with dynamic three-dimensional MR renography. Radiology 270(2):409–415. https://doi.org/10.1148/radiol.13122495
Liu X, Murphy MP, Xing W, Wu H, Zhang R, Sun H (2018) Mitochondria-targeted antioxidant MitoQ reduced renal damage caused by ischemia-reperfusion injury in rodent kidneys: longitudinal observations of T. Magn Reson Med 79(3):1559–1567. https://doi.org/10.1002/mrm.26772
Sari-Sarraf F, Pomposiello S, Laurent D (2008) Acute impairment of rat renal function by L -NAME as measured using dynamic MRI. MAGMA 21(4):291–297. https://doi.org/10.1007/s10334-008-0130-6
Privratsky JR, Wang N, Qi Y, Ren J, Morris BT, Hunting JC, Johnson GA, Crowley SD (2019) Dynamic contrast-enhanced MRI promotes early detection of toxin-induced acute kidney injury. Am J Physiol Renal Physiol 316(2):F351–F359. https://doi.org/10.1152/ajprenal.00416.2018
Jiang K, Tang H, Mishra PK, Macura SI, Lerman LO (2017) Measurement of murine single-kidney glomerular filtration rate using dynamic contrast-enhanced MRI. Magn Reson Med 79(6):2935–2943. https://doi.org/10.1002/mrm.26955
Zollner FG, Zimmer F, Klotz S, Hoeger S, Schad LR (2014) Renal perfusion in acute kidney injury with DCE-MRI: deconvolution analysis versus two-compartment filtration model. Magn Reson Imaging 32(6):781–785. https://doi.org/10.1016/j.mri.2014.02.014
Oostendorp M, de Vries EE, Slenter JM, Peutz-Kootstra CJ, Snoeijs MG, Post MJ, van Heurn LW, Backes WH (2011) MRI of renal oxygenation and function after normothermic ischemia-reperfusion injury. NMR Biomed 24(2):194–200. https://doi.org/10.1002/nbm.1572
Zöllner FG, Zimmer F, Klotz S, Hoeger S, Schad LR (2015) Functional imaging of acute kidney injury at 3 tesla: investigating multiple parameters using DCE-MRI and a two-compartment filtration model. Z Med Phys 25(1):58–65. https://doi.org/10.1016/j.zemedi.2014.01.002
Notohamiprodjo M, Kalnins A, Andrassy M, Kolb M, Ehle B, Mueller S, Thomas MN, Werner J, Guba M, Nikolaou K, Andrassy J (2016) Multiparametric functional MRI: a tool to uncover subtle changes following allogeneic renal transplantation. PLoS One 11(11):e0165532. https://doi.org/10.1371/journal.pone.0165532
Acknowledgments
The Italian Ministry for Education and Research (MIUR) is gratefully acknowledged for yearly FOE funding to the Euro-BioImaging Multi-Modal Molecular Imaging Italian Node (MMMI).
This publication is based upon work from COST Action PARENCHIMA, supported by European Cooperation in Science and Technology (COST). COST (www.cost.eu) is a funding agency for research and innovation networks. COST Actions help connect research initiatives across Europe and enable scientists to enrich their ideas by sharing them with their peers. This boosts their research, career, and innovation.
PARENCHIMA (renalmri.org) is a community-driven Action in the COST program of the European Union, which unites more than 200 experts in renal MRI from 30 countries with the aim to improve the reproducibility and standardization of renal MRI biomarkers.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this protocol
Cite this protocol
Pedersen, M. et al. (2021). Dynamic Contrast Enhancement (DCE) MRI–Derived Renal Perfusion and Filtration: Basic Concepts. In: Pohlmann, A., Niendorf, T. (eds) Preclinical MRI of the Kidney. Methods in Molecular Biology, vol 2216. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0978-1_12
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0978-1_12
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0977-4
Online ISBN: 978-1-0716-0978-1
eBook Packages: Springer Protocols