Abstract
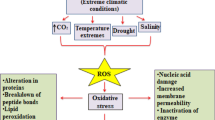
The REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 (RRTF1) gene encodes a member of the ERF/AP2 transcription factor family involved in redox homeostasis. The RRTF1 gene shows tissue-specific responsiveness to various abiotic stress treatments including a response to salt stress in roots. An interesting feature of this response is an adaptation phase that follows its activation, when promoter levels revert to a base line level, even if salt stress is maintained. It is unclear if adaption is controlled by a switch in promoter activity or by changes in transcript levels. Here we show that the RRTF1 promoter is sufficient for the control of both activation and adaptation to salt stress. As constitutive expression of RRTF1 turned out to be detrimental to the plant, we propose that promoter-regulated adaptation evolved as a protection mechanism to balance the beneficial effects of short-term gene activation and the detrimental effects of long-term gene expression.




Similar content being viewed by others
References
Shahmuradov, I. A., Gammerman, A. J., Hancock, J. M., Bramley, P. M., & Solovyev, V. V. (2003). PlantProm: A database of plant promoter sequences. Nucleic Acids Research, 31, 114–117.
Isabelle, B. S., Stéphanie, P., Françoise, C., Juliette, L., & Christophe, B. (2017). 5′ to 3′ mRNA decay contributes to the regulation of arabidopsis seed germination by dormancy. Plant Physiology, 173(3), 1709–1723.
Middleton, A. M., Úbeda-Tomás, S., Griffiths, J., Holman, T., Hedden, P., et al. (2012) Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling. Proceedings of the National Academy of Sciences, 109, 7571–7576.
Borsani, O., Zhu, J., Verslues, P. E., Sunkar, R., & Zhu, J.K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in arabidopsis. Cell, 123, 1279–1291.
Lämke, J., & Bäurle, I. (2017). Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biology, 18, 124.
Khandelwal, A., Elvitigala, T., Ghosh, B., & Quatrano, R. S. (2008). Arabidopsis transcriptome reveals control circuits regulating redox homeostasis and the role of an AP2 transcription factor. Plant Physiology, 148, 2050–2058.
Jen, C.-H., Michalopoulos, I., Westhead, D., & Meyer, P. (2005). Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biology, 6, R51.
Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., & Gruissem, W. (2004). GENEVESTIGATOR. arabidopsis microarray database and analysis toolbox. Plant Physiology, 136, 2621–2632.
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal, 50, 347–363.
Fang, Y.Y., Zhao, J.H., Liu, S.W., Wang, S., Duan, C.G., & Guo, H.S. (2016). CMV2b-AGO interaction is required for the suppression of RDR-dependent antiviral silencing in Arabidopsis. Frontiers in Microbiology, 7, 1329.
McHale, M., Eamens, A. L., Finnegan, E. J., & Waterhouse, P. M. (2013). A 22-nt artificial micro-RNAmediates widespread RNA silencing in Arabidopsis. The Plant Journal, 76(3), 519–529.
Katiyar-Agarwal, S., Morgan, R., Dahlbeck, D., Borsani, O., Villegas, A., et al. (2006) A pathogen-inducible endogenous siRNA in plant immunity. Proceedings of the National Academy of Sciences, 103, 18002–18007.
Hu, Y., Chen, L., Wang, H., Zhang, L., Wang, F., & Yu, D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. The Plant Journal, 74, 730–745.
Karan, R., DeLeon, T., Biradar, H., & Subudhi, P. K. (2012). Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS ONE, 7, e40203.
Pandey, G., Sharma, N., Sahu, P. P., & Prasad, M. (2016). Chromatin-based epigenetic regulation of plant abiotic stress response. Current Genomics, 17(6), 490–498.
Zubko, E., Gentry, M., Kunova, A., & Meyer, P. (2012). De novo DNA methylation activity of METHYLTRANSFERASE 1 (MET1) partially restores body methylation in Arabidopsis thaliana. The Plant Journal, 71, 1029–1037.
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L., & Poethig, R. S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes & Development, 18, 2368–2379.
Kankel, M. W., Ramsey, D. E., Stokes, T. L., Flowers, S. K., Haag, J. R., Jeddeloh, J. A., et al. (2003). Arabidopsis MET1 cytosine methyltransferase mutants. Genetics, 163, 1109–1122.
Adamo, A. (2008). Characterisation of unusual transcript classes. In Arabidopsis thaliana (196 p). Leeds: University of Leeds.
Gallois, J.-L., Woodward, C., Reddy, G. V., & Sablowski, R. (2002). Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development, 129, 3207–3217.
Clough, S. J., & Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal, 16, 735–743.
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., et al. (2007). The At Gen Express global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant Journal, 50, 347–363.
Livak, K., & Schmittgen, T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 −∆∆CT method. Methods, 25, 402–408.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Soliman, E.R.S., Meyer, P. Responsiveness and Adaptation to Salt Stress of the REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 (RRTF1) Gene are Controlled by its Promoter. Mol Biotechnol 61, 254–260 (2019). https://doi.org/10.1007/s12033-019-00155-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-019-00155-9