Abstract
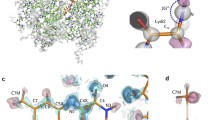
The 3D full-atom model of the whole-size CYP102A1 from Bacillus megaterium (cytochrome P450 BM3) has been constructed using molecular modeling methods. The structure model was constructed using crystal structures of the separate FAD-binding domain (PDB ID: 4DQK) and the complex of FMN-binding and monooxygenase domains (PDB ID: 1BVY). Modeling procedure included analysis of the domains’ surfaces to find the orientation with maximum inter-subunit contacts. The overall configuration of the obtained complex was optimized using molecular dynamics. The final full-atom structure model shows rather tight interactions between FAD- and FMN-binding domains due to 10 inter-domain hydrogen bonds and hydrophobic interactions between three pairs of amino acid residues. This 3D model can be used for structure-function studies and rational design of the enzyme as well as for construction of hybrid supramolecular structures of biocatalysts with cytochrome P450 BM3.



Similar content being viewed by others
REFERENCES
Neeli, R. Girvan, H.M., Lawrence, A., Warren, M.J., Leys, D., Scrutton, N.S., and Munro, A.W., FEBS Lett., 2005, vol. 579, p. 5582.
Girvan, H.M. and Munro, A.W., Curr. Opin. Chem. Biol., 2016, vol. 31, p. 136.
Joyce, M.G., Ekanem, I.S., Roitel, O., Dunford, A.J., Neeli, R., Girvan, H.M., Baker, G.J., Curtis, R.A., Munro A.W., and Leys, D., FEBS J., 2012, vol. 279, p. 1694.
Sevrioukova, I.F., Li, H., Zhang, H., Peterson, J.A., and Poulos, T.L., Proc. Natl. Acad. Sci. U. S. A., 1999, vol. 96, p. 1863.
Bridges, A., Gruenke, L., Chang, Y. T., Vakser, I. A., Loew, G., and Waskell, L., J. Biol. Chem., 1998, vol. 273, p. 17036.
Shimizu, T., Tateishi, T., and Fujii-Kuriyama, H.M., J. Biol. Chem., 1991, vol. 266, p. 3372.
Stayton P.S., and Sligar, S. G., Biochemistry, 1991, vol. 30, p. 7381.
Wada, A., and Waterman, M.R., J. Biol. Chem., 1992, vol. 267, p. 22877.
Roccatano, D., J. Phys.: Condens. Matter, 2015, vol. 27, 273102.
Guengerich, F.P., Michael, R., Waterman, M.R., and Egli, M., Trends Pharmacol. Sci., 2016, vol. 37, p. 625.
Ludwig, M.L., Pattridge, K.A., Metzger, A.L., Di-xon, M.M., Eren, M., Feng, Y., and Swenson, R.P., Biochemistry, 1997, vol. 36, p. 1259.
Nelson, M.T., Humphrey, W.F. Gursoy, A., Dalke, A., Kalé, L.V., Skeel, R.D., and Schulten, K., Int. J. Supercomput. Appl. High Perform. Comput., 1996, vol. 10, no. 4, p. 251.
Humphrey, W., Dalke, and A., Schulten, K., J. Mol. Graphics Modell., 1996, vol. 14, p. 33.
Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M., J. Comput. Chem., 1983, vol. 4, no. 2, p. 187.
MacKerell, A.D., Jr., Bashford, D., Bellott, M., Dunbrack, R.L., Jr., Evanseck, J.D., Field, M.J., Fischer, S., Gao, J., Guo, H., Ha, S., McCarthy, J.D., Kuchnir, L., Kuczera, K., Lau, F.T.K., Mattos, C., Michnick, S., Ngo, T., Nguyen, D.T., Prodhom, B., Reiher, W.E., Roux, B., Schlenkrich, M., Smith, J.C., Stote, R., Straub, J., Watanabe, M., Wiórkiewicz-Kuczera, J., Yin, D., and Karplus, M., J. Phys. Chem., 1998, vol. 102, p. 3586.
Vanommeslaeghe, K., Hatcher, E., and Acharya, C., J. Comput. Chem., 2010, vol. 31, p. 671.
Tishkov, V.I., and Khoronenkova, S.V., Biochemistry (Moscow), 2005, vol. 70, no. 1, p. 40.https://doi.org/10.1007/s10541-005-0050-2
Tishkov, V.I., Khoronenkova, S.V., Cherskova, N.V., Savin, S.S., Uporov, I.V., Moscow Univ. Chem. Bull. (Engl. Transl.), 2010, vol. 65, no. 3, p. 121.https://doi.org/10.3103/S0027131410030028
Cherskova, N.V., Khoronenkova, S.V., and Tishkov, V.I., Russ. Chem. Bull., 2010, vol. 59, no. 1, p. 269.https://doi.org/10.1007/s11172-010-0072-9
Voevodin, Vl., Antonov, A., Nikitenko, D., Shvets, P., Sobolev, S., Sidorov, I., Stefanov, K., Voevodin, Vad., and Zhumatiy S., Supercomput. Front. Innovations, 2019, vol. 6, no. 2, p. 4.
ACKNOWLEDGMENTS
This work was performed using the equipment of the shared research facilities of HPC computing resources at the Moscow State University [20].
Funding
This work was partially supported by the Russian Science Foundation (project no. 18-74-00146).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Supplementary material
There are no additional materials.
About this article
Cite this article
Krivitskaya, A.V., Pometun, A.A., Parshin, P.D. et al. Full Structure Modeling of Three-Domains Monooxygenase CYP102A1 ВМ3 from Bacillus megaterium. Moscow Univ. Chem. Bull. 75, 162–166 (2020). https://doi.org/10.3103/S0027131420030074
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0027131420030074