Abstract
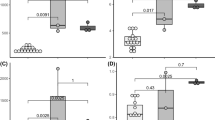
The bacterial diversity of Pectinatella magnifica colonies sampled from pounds in South Bohemia during the summer of 2012 was investigated. The bacterial counts determined after cultivation on modified yeast extract-tryptone agar (Oxoid) supplemented with glucose (1 g L−1) varied from 4.22 to 6.61 and from 1.30 to 6.85 log CFU/g for aerobes and anaerobes, respectively. Higher counts were found in the superficial structures of Pectinatella colonies than in the inner gelled mass. Neither a trend in bacterial numbers at the individual site during the season, nor correlations between bacterial counts in P. magnifica and the surrounding water were observed. Fifty-four isolates were identified by sequencing the 16S rRNA gene and through MALDI-TOF MS analysis. Species of Aeromonas and Aquitalea were the predominantly isolated bacteria, but members of Chryseobacterium, Herbaspirillum, Enterobacter, Lactococcus, Leuconostoc, Pseudomonas and Sphingomonas were also found. As listed genera are wildly distributed in different water, soil, and plant samples, we conclude that Pectinatella colonies are inhabited by environmental bacteria. Nevertheless, a symbiotic relationship of these bacteria with P. magnifica cannot be excluded.
Similar content being viewed by others
References
Anderson C.M. & Haygood M.G. 2007. α-Protobacterial symbionts of marine bryozoans in the genus Watersipora. Appl. Environ. Microbiol. 73: 303–311. DOI: 10.1128/AEM.00604-06
Balounová Z., Rajchard J., Švehla J. & Šmahel L. 2011. The onset of invasion of bryozoan Pectinatella magnifica in South Bohemia (Czech Republic). Biologia 66: 1091–1096. DOI: 10.2478/s11756-011-0118-y
Bernauer D. & Jansen W. 2006. Recent invasions of alien macroinvertebrates and loss of native species in upper Rhine River, Germany. Aquatic Invasions 2: 55–71. DOI 10.3391/ai.2006.1.2.2
Canning E.U., Refardt D., Vossbrinck Ch.R., Okamura B. & Curry A. 2002. New diplokaryotic microsporidia (Phylum Microsporidia) from freshwater bryozoans (Bryozoa, Phylacto-laemata). Eur. J. Protistol. 38: 247–265. DOI: 10.1078/0932-4739-00867
Denner E.B.M., Paukner S., Kämpfer P., Moore E.R.B., Abraham W.R., Busse H.J., Wanner G. & Lubitz W. 2001. Sphingomonas pituitosa sp. nov., an exopolysaccharide-producing bacterium that secretes an unusual type of sphingan. Int. J. Syst. Evol. Microbiol. 51: 827–841. DOI: 10.1099/00207713-51-3-827
Ding L. & Yokota A. 2004. Proposals of Curvibacter gracilis gen nov., sp. nov. and Herbaspirillum putei sp. nov. for bacterial strains isolated from well water and reclassification of [Pseudomonas] huttiensis, [Pseudomonas] lanceolata, [Aquaspirillum] delicatum and [Aquaspirillum]autotrophicum as Herbaspirillum huttiense comb. nov., Curvibacter lanceolatus comb. nov., Curvibacter delicatus comb. nov. and Herbaspirillum autotrophicum comb. nov. Int. J. Syst. Evol. Microbiol. 54: 2223–2230. DOI: 10.1099/ijs.0.02975-0
Dobritsa A.P., Reddy M.C.S. & Samadpour M. 2010. Reclassification of Herbaspirillum putei as a later heterotypic synonym of Herbaspirillum huttiense, with the description of H. huttiense subsp. huttiense subsp. nov. and H. huttiense subsp. putei subsp. nov., and description of Herbaspirillumaquaticum sp. nov. Int. J. Syst. Evol. Microbiol. 60: 1418–1426. DOI: 10.1099/ijs.0.009381-0
Forster R.J., Teather R.M., Gong J. & Deng S.J. 1995. 16S rRNA analysis of Butyrivibrio fibrisolvens: phylogenetic position and relation to butyrate producing anaerobic bacteria from the rumen of a shorttailed deer. Lett. Appl. Microbiol. 23: 218–222. PMID: 8987694
González J.M., Whitman W.B., Hodson R.E. & Moran M.A. 1996. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl. Environ. Microbiol. 62: 4433–4440. PM-CID: PMC168269
Grimont P.A.D. & Grimont F. 2005. Enterobacter. pp. 661–669. In: Brenner D.J., Krieg N.R. & Staley J.T. (eds), Bergey’s Manual of Systematic Bacteriology, Springer, New York, USA.
Hall T.A. 1999. BioEdit: a userfriendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41: 95–98.
Herzog P., Winkler I., Wolking D., Kämpfer P. & Lipski A. 2008. Chryseobacterium ureilyticum sp. nov., Chryseobacterium gambrini sp. nov., Chryseobacterium pallidum sp. nov. and Chryseobacterium molle sp. nov., isolated from beerbottling plants. Int. J. Syst. Evol. Microbiol. 58: 26–33. DOI: 10.1099/ijs.0.65362-0
Holzapfel W.H., Björkroth J.A. & Dicks L.M.T. 2009. Leuconostoc. pp. 624–635. In: De Vos P., Garrity G.M., Jones D., Krieg N.R., Ludwig W., Rainey F.A., Schleifer K.H. & Whitman W.B. (eds), Bergey’s Manual of Systematic Bacteriology, Vol 3, The Firmicutes, Springer, New York, USA, 1450 pp. DOI: 10.1007/978-0-387-68489-5, ISBN: 978-0-387-95041-9
Indergand S. & Graf J. 2000. Ingested blood contributes to the specificity of the symbiosis of Aeromonas veronii biovar so-bria and Hirudo medicinalis, the medicinal leech. Appl. Environ. Microbiol. 66: 4735–4741. PMID: 11055917
Joo G.J., Ward A.K. & Ward G.M. 1992. Ecology of Pectinatella magnifica (Bryozoa) in an Alabama Oxbow lake: Colony growth and association with algae. J. N. Am. Benthol. Soc. 11: 324–333.
Kämpfer P., Chandel K., Prasad G.B.K.S., Shouche Y.S. & Veer V. 2010. Chryseobacterium culicis sp. nov., isolated from the midgut of the mosquito Culex quinquefasciatus. Int. J. Syst. Evol. Microbiol. 60: 2387–2391. DOI: 10.1099/ijs.0.019794-0
Kmeť V. & Drugdová Z. 2012. Antimicrobial susceptibility of microflora from ovine cheese. Folia Microbiol. 57: 291–293. DOI: 10.1007/s12223-012-0128-3
Lau H.T., Faryna J. & Triplett E.W. 2006. Aquitalea magnusonii gen. nov., sp. nov., a novel Gram-negative bacterium isolated from a humic lake. Int. J. Syst. Evol. Microbiol. 56: 867–871. DOI: 10.1099/ijs.0.64089-0
Lee C.M., Weon H.Y., Kim Y.J., Son J.A., Yoon S.H., Koo B.S. & Kwon S.W. 2009. Aquitalea denitrificans sp. nov., isolated from a Korean wetland. Int. J. Syst. Evol. Microbiol. 59: 1045–1048. DOI: 10.1099/ijs.0.002840-0
Lim G.E. & Haygood M.G. 2004. ‘Candidatus Endobugula glebosa’ a specific bacterial symbiont of the marine bryozoan Bugula simplex. Appl. Environ. Microbiol. 70: 4921–4929. DOI: 10.1128/AEM.70.8.4921-4929.2004
Lim-Fong G.E., Regali L.A. & Haygood M.G. 2008. Evolutionary relationships of ‘Candidatus Endobugula’ bacterial symbionts and their Bugula bryozoan hosts. Appl. Environ. Microbiol. 74: 3605–3609. DOI: 10.1128/AEM.02798-07
Maidak B.L., Larsen N., McCaughey M.J., Overbeek R., Olsen G.J., Forgel K., Blandy J. & Woese C.R. 1994. The ribosomal database project. Nucl. Acids Res. 22: 3485–3487. DOI: 10.1093/nar/24.1.82
Opravilová V. 2006. Bryozoa–mechovky, p. 366. In: Mlíkovský J. & Stýblo P. (eds), Nepůvodní druhy fauny a flóry České republiky, ČSOP, Praha, CZ, 496 pp. ISBN: 80-86770-17-6
Pukall R., Kramer I., Rohde M. & Stackebrandt E. 2001. Microbial diversity of cultivatable bacteria associated with the North Sea bryozoan Flustra foliacea. Syst. Appl. Microbiol. 24: 623–633. DOI: 10.1078/0723-2020-00073
Rodriguez S. & Vergon J.P. 2002. Pectinatella magnifica Leidy 1851 (Phylactolaemates), a species of Bryozoa introduced in the north of Franche-Comte. Bull. Fr. P˛eche Piscic. 365–366: 281–296.
Silver A.C., Williams D., Faucher J., Horneman A.J., Gogarten J.P. & Graf J. 2011. Complex evolutionary history of Aeromonas veronii group revealed by host interaction and DNA sequence data. Plos One 6:e16751. DOI: 10.1371/jour-nal.pone.0016751
Teuber M. 2009. Lactococcus. pp. 711–722. In: De Vos P., Garrity G.M., Jones D., Krieg N.R., Ludwig W., Rainey F.A., Schleifer K.H. & Whitman W.B. (eds), Bergey’s Manual of Systematic Bacteriology, Vol 3, The Firmicutes, Springer, New York, USA, 1450 pp. DOI: 10.1007/978-0-387-68489-5, ISBN: 978-0-387-95041-9
Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 24: 4876–4882. PMID: 9396791
Tvrzová L., Schumann P., Spröer C., Sedláček I., Páčová Z., Šedo O., Zdráhal Z., Steffen M. & Lang E. 2006. Pseudomonas moraviensis sp. nov. and Pseudomonas vranovensis sp. nov., soil bacteria isolated on nitroaromatic compounds, and emended description of Pseudomonas asplenii. Int. J. Syst. Evol. Microbiol. 56: 2657–2663. DOI: 10.1099/ijs.0.63988-0
Valverde A., Vlázquez E., Gutiérrez C., Cervantes E., Ventosa A. & Igual J.M. 2003. Herbaspirillum lusitanum sp. nov., a novel nitrogen-fixing bacterium associated with root nodules of Phaseolus vulgaris. Int. J. Syst. Evol. Microbiol. 53: 1979–1983. DOI: 10.1099/ijs.0.02677-0
Weisburg W.G., Barns S.M., Pelletier D.A. & Lane D.J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173: 697–703.
Acknowledgements
This study was supported by a grant GACR P503/12/0337 of the Czech Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vlková, E., Killer, J., Kmeť, V. et al. Identification of microbiota associated with Pectinatella magnifica in South Bohemia. Biologia 70, 365–371 (2015). https://doi.org/10.1515/biolog-2015-0040
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1515/biolog-2015-0040