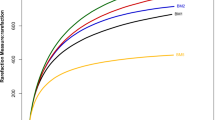
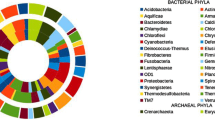
Abstract
The mangrove ecosystem is the world’s fourth most productive ecosystem in terms of service value and offering rich biological resources. Microorganisms play vital roles in these ecological processes, thus researching the mangroves-microbiota is crucial for a deeper comprehension of mangroves dynamics. Amplicon sequencing that targeted V4 region of 16S rRNA gene was employed to profile the microbial diversities and community compositions of 19 soil samples, which were collected from the rhizosphere of 3 plant species (i.e., Avicennia marina, Ceriops tagal, and Rhizophora mucronata) in the mangrove forests of Lasbela coast, Pakistan. A total of 67 bacterial phyla were observed from three mangroves species, and these taxa were classified into 188 classes, 453 orders, 759 families, and 1327 genera. We found that Proteobacteria (34.9–38.4%) and Desulfobacteria (7.6–10.0%) were the dominant phyla followed by Chloroflexi (6.6–7.3%), Gemmatimonadota (5.4–6.8%), Bacteroidota (4.3–5.5%), Planctomycetota (4.4–4.9%) and Acidobacteriota (2.7–3.4%), Actinobacteriota (2.5–3.3%), and Crenarchaeota (2.5–3.3%). After considering the distribution of taxonomic groups, we prescribe that the distinctions in bacterial community composition and diversity are ascribed to the changes in physicochemical attributes of the soil samples (i.e., electrical conductivity (ECe), pH, total organic matter (OM), total organic carbon (OC), available phosphorus (P), and extractable potassium (CaCO3). The findings of this study indicated a high-level species diversity in Pakistani mangroves. The outcomes may also aid in the development of effective conservation policies for mangrove ecosystems, which have been hotspots for anthropogenic impacts in Pakistan. To our knowledge, this is the first microbial research from a Pakistani mangrove forest.





Similar content being viewed by others
Data availability
The collected reads were deposited in GenBank under Bio Project accession number PRJNA817083 and BioSample accession numbers SAMN26747204–SAMN26747222 (in total 19 samples).
References
Alongi DM (2012) Carbon sequestration in mangrove forests. Carbon Manag 3:313–322
Alongi DM (2020) Nitrogen cycling and mass balance in the world’s mangrove forests. Nitrogen 1:167–189
Ananda K, Sridhar K (2002) Diversity of endophytic fungi in the roots of mangrove species on the west coast of India. Can J Microbiol 48:871–878
Andreote FD, Jimenez DJ, Chaves D, Dias AC, Luvizotto DM, Dini-Andreote F, Fasanella CC, Lopez MV, Baena S, Taketani RG, de Melo IS (2012) The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS ONE 7:e38600. https://doi.org/10.1371/journal.pone.0038600
Aziz I, Khan MA (2000) Physiological adaptations of Avicennia marina to seawater concentrations in the indus delta, Pakistan. Pak J Bot 32:151–169
Azman AS, Othman I, Velu SS, Chan KG, Lee LH (2015) Mangrove rare actinobacteria: taxonomy, natural compound, and discovery of bioactivity. Front Microbiol 6:856. https://doi.org/10.3389/fmicb.2015.00856
Balk M, Keuskamp JA, Laanbroek HJ (2016) Potential for sulfate reduction in mangrove forest soils: comparison between two dominant species of the Americas. Front Microbiol 7:1855. https://doi.org/10.3389/fmicb.2016.01855
Basak P, Pramanik A, Roy R, Chattopadhyay D, Bhattacharyya M (2015) Cataloguing the bacterial diversity of the Sundarbans mangrove, India in the light of metagenomics. Genom Data 4:90–92. https://doi.org/10.1016/j.gdata.2015.03.014
Basyuni M, Slamet B, Sulistiyono N, Munir E, Vovides A, Bunting P (2021) Physicochemical characteristics, nutrients, and fish production in different types of mangrove forests in North Sumatra and the Aceh Provinces of Indonesia. Kuwait J Sci 48
Bianchi TS (2011) The role of terrestrially derived organic carbon in the coastal ocean: a changing paradigm and the priming effect. Proc Nat Acad Sci 108:19473–19481
Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH (2008) Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci 105:10583–10588. https://doi.org/10.1073/pnas.0709942105
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodriguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vazquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Bukin YS, Galachyants YP, Morozov I, Bukin S, Zakharenko A, Zemskaya T (2019) The effect of 16S rRNA region choice on bacterial community metabarcoding results. Sci Data 6:1–14
Cabral L, Júnior GVL, de Sousa STP, Dias ACF, Cadete LL, Andreote FD, Hess M, de Oliveira VM (2016) Anthropogenic impact on mangrove sediments triggers differential responses in the heavy metals and antibiotic resistomes of microbial communities. Environ Pollut 216:460–469
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108(Suppl 1):4516–4522. https://doi.org/10.1073/pnas.1000080107
Carlson RR, Evans LJ, Foo SA, Grady BW, Li J, Seeley M, Xu Y, Asner GP (2021) Synergistic benefits of conserving land-sea ecosystems. Glob Ecol Conserv 28:e01684
Curtis TP, Sloan WT (2004) Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr Opin Microbiol 7:221–226. https://doi.org/10.1016/j.mib.2004.04.010
Das S, De M, Ganguly D, Maiti TK, Mukherjee A, Jana TK, De TK (2012) Depth integrated microbial community and physico-chemical properties in mangrove soil of Sundarban. India Adv Microbiol 2:234
Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, Maestre FT, Singh BK, Fierer N (2018) A global atlas of the dominant bacteria found in soil. Sci 359:320–325
Díez-Vives C, Gasol JM, Acinas SG (2014) Spatial and temporal variability among marine Bacteroidetes populations in the NW Mediterranean Sea. Syst Appl Microbiol 37:68–78
dos Santos HF, Cury JC, do Carmo FL, dos Santos AL, Tiedje J, van Elsas JD, Rosado AS, Peixoto RS (2011) Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: bacterial proxies for oil pollution. PLoS ONE 6:e16943. https://doi.org/10.1371/journal.pone.0016943
Dubey A, Malla MA, Khan F, Chowdhary K, Yadav S, Kumar A, Sharma S, Khare PK, Khan ML (2019) Soil microbiome: a key player for conservation of soil health under changing climate. Biodivers Conserv 28:2405–2429
Edgar CR (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–1001. https://doi.org/10.1038/nmeth.2604
Fadeev E, Cardozo-Mino MG, Rapp JZ, Bienhold C, Salter I, Salman-Carvalho V, Molari M, Tegetmeyer HE, Buttigieg PL, Boetius A (2021) Comparison of two 16S rRNA primers (V3–V4 and V4–V5) for studies of arctic microbial communities. Front Microbiol 12:637526
Fernandes SO, Kirchman DL, Michotey VD, Bonin PC, LokaBharathi PA (2014) Bacterial diversity in relatively pristine and anthropogenically-influenced mangrove ecosystems (Goa, India). Braz J Microbiol 45:1161–1171. https://doi.org/10.1590/s1517-83822014000400006
Foesel BU, Rohde M, Overmann J (2013) Blastocatella fastidiosa gen. nov., sp. nov., isolated from semiarid savanna soil - the first described species of Acidobacteria subdivision 4. Syst Appl Microbiol 36:82–89. https://doi.org/10.1016/j.syapm.2012.11.002
Fortunato CS, Crump BC (2015) Microbial gene abundance and expression patterns across a river to ocean salinity gradient. PLoS ONE 10:e0140578
Ghizelini AM, Mendonça-Hagler LCS, Macrae A (2012) Microbial diversity in Brazilian mangrove sediments: a mini review. Brazil J Microbiol 43:1242–1254
Ghosh A, Bhadury P (2018) Investigating monsoon and post-monsoon variabilities of bacterioplankton communities in a mangrove ecosystem. Environ Sci Pollut Res 25:5722–5739
Ghosh A, Dey N, Bera A, Tiwari A, Sathyaniranjan KB, Chakrabarti K, Chattopadhyay D (2010) Culture independent molecular analysis of bacterial communities in the mangrove sediment of Sundarban. India Saline Syst 6:1. https://doi.org/10.1186/1746-1448-6-1
Gilbert JA, Meyer F, Bailey MJ (2011) The future of microbial metagenomics (or is ignorance bliss?). ISME J 5:777–779
Giri C, Ochieng E, Tieszen LL, Zhu Z, Singh A, Loveland T, Masek J, Duke N (2011) Status and distribution of mangrove forests of the world using earth observation satellite data. Glob Ecol Biogeogr 20:154–159. https://doi.org/10.1111/j.1466-8238.2010.00584
Gomes NC, Flocco CG, Costa R, Junca H, Vilchez R, Pieper DH, Krogerrecklenfort E, Paranhos R, Mendonca-Hagler LC, Smalla K (2010) Mangrove microniches determine the structural and functional diversity of enriched petroleum hydrocarbon-degrading consortia. FEMS Microbiol Ecol 74:276–290. https://doi.org/10.1111/j.1574-6941.2010.00962.x
Gotelli NJ, Chao A (2013) Measuring and estimating species richness, species diversity, and biotic similarity from sampling data. In: Levin SA (ed) Encyclopedia of Biodiversity, 2nd edn. Academic Press, Waltham, pp 195–211
Haldar S, Nazareth SW (2018) Taxonomic diversity of bacteria from mangrove sediments of Goa: metagenomic and functional analysis. 3 Biotech 8:1–10
Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579
Hewson I, Steele JA, Capone DG, Fuhrman JA (2006) Remarkable heterogeneity in meso-and bathypelagic bacterioplankton assemblage composition. Limnol Oceanogr 51:1274–1283
Holguin G, Vazquez P, Bashan Y (2001) The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol Fert Soils 33:265–278. https://doi.org/10.1007/s003740000319
Hong Y, Liao D, Hu A, Wang H, Chen J, Khan S, Su J, Li H (2015) Diversity of endophytic and rhizoplane bacterial communities associated with exotic Spartina alterniflora and native mangrove using Illumina amplicon sequencing. Canad J Microbiol 61:723–733
Hou L, Zheng Y, Liu M, Gong J, Zhang X, Yin G, You L (2013) Anaerobic ammonium oxidation (anammox) bacterial diversity, abundance, and activity in marsh sediments of the Yangtze Estuary. J Geophys Res Biogeosci 118:1237–1246
Hu C, Cao Z (2007) Size and activity of the soil microbial biomass and soil enzyme activity in long-term field experiments. World J Agr Sci 3:63–70
Imchen M, Kumavath R, Barh D, Vaz A, Góes-Neto A, Tiwari S, Ghosh P, Wattam AR, Azevedo V (2018) Comparative mangrove metagenome reveals global prevalence of heavy metals and antibiotic resistome across different ecosystems. Sci Rep 8:1–15
Jansson JK, Hofmockel KS (2020) Soil microbiomes and climate change. Nat Rev Microbiol 18:35–46
Jiang XT, Peng X, Deng GH, Sheng HF, Wang Y, Zhou HW, Tam NF (2013) Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb Ecol 66:96–104. https://doi.org/10.1007/s00248-013-0238-8
Joshi H, Ghose M (2003) Forest structure and species distribution along soil salinity and pH gradient in mangrove swamps of the Sundarbans. Trop Ecol 44:195–204
Kannan S, Balamurugan S, Ragavan P, Deivasigamani B, Wee A, Salmo S, Basyuni M, Kajita T (2022) eDNA envisaged conservation of IUCN threatened taxa of the tropical mangrove ecosystems. IOP Conf Ser Earth Environ Sci 1115:012032 (IOP Publishing)
Kathiresan K, Bingham BL (2001) Biology of mangroves and mangrove ecosystems. Adv Mar Biol 40:81–251. https://doi.org/10.1016/S0065-2881(01)40003-4
Kersters K, De Vos P, Gillis M, Swings J, Vandamme P, Stackebrandt E (2006) Introduction to the proteobacteria. The prokaryotes: a handbook on the biology of bacteria. Springer, pp 3–37
Kim M, Morrison M, Yu Z (2011) Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J Microbiol Methods 84:81–87
Kirchman DL, Dittel AI, Malmstrom RR, Cottrell MT (2005) Biogeography of major bacterial groups in the Delaware Estuary. Limnol Oceanogr 50:1697–1706
Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R (2012) Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol 27:1E.5.1–1E.5.20
Li M, Cao H, Hong Y, Gu JD (2011) Spatial distribution and abundances of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in mangrove sediments. Appl Microbiol Biotechnol 89:1243–1254. https://doi.org/10.1007/s00253-010-2929-0
Liang J-B, Chen Y-Q, Lan C-Y, Tam NFY, Zan Q-J, Huang L-N (2006) Recovery of novel bacterial diversity from mangrove sediment. Mar Biol 739–747. https://doi.org/10.1007/s00227-006-0377-2
Liao P-C, Huang B-H, Huang S (2007) Microbial community composition of the Danshui river estuary of Northern Taiwan and the practicality of the phylogenetic method in microbial barcoding. Microbial Ecol 54:497–507
Liao S, Wang Y, Liu H, Fan G, Sahu SK, Jin T, Chen J, Zhang P, Gram L, Strube ML (2020) Deciphering the microbial taxonomy and functionality of two diverse mangrove ecosystems and their potential abilities to produce bioactive compounds. Msystems 5:e00851-e1819
Liu M, Huang H, Bao S, Tong Y (2019) Microbial community structure of soils in Bamenwan mangrove wetland. Sci Rep 9:8406. https://doi.org/10.1038/s41598-019-44788-x
Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, Aklujkar M, Butler JE, Giloteaux L, Rotaru AE, Holmes DE, Franks AE, Orellana R, Risso C, Nevin KP (2011) Geobacter: the microbe electric’s physiology, ecology, and practical applications. Adv Microb Physiol 59:1–100. https://doi.org/10.1016/B978-0-12-387661-4.00004-5
Lyimo TJ, Pol A, Harhangi HR, Jetten MS, Op den Camp HJ (2009) Anaerobic oxidation of dimethylsulfide and methanethiol in mangrove sediments is dominated by sulfate-reducing bacteria. FEMS Microbiol Ecol 70:483–492
Lyimo TJ, Pol A, den Camp HJO (2002) Sulfate reduction and methanogenesis in sediments of Mtoni mangrove forest, Tanzania. AMBIO J Hum Environ 31:614–616
Menéndez P, Losada IJ, Torres-Ortega S, Narayan S, Beck MW (2020) The global flood protection benefits of mangroves. Sci Rep 10:4404. https://doi.org/10.1038/s41598-020-61136-6
Miller RO, Gavlak R, Horneck D (2013) Soil, plant and water reference methods for the western region. Fort Collins, CO, USA, Colorado State University
Mujakić I, Piwosz K, Koblížek M (2022) Phylum Gemmatimonadota and its role in the environment. Microorganisms 10:151
Nguyen LD, Nguyen CT, Le HS, Tran BQ (2019) Mangrove mapping and above-ground biomass change detection using satellite images in coastal areas of Thai Binh Province. Vietnam for Soc 3:248–261
Nogales B, Aguiló-Ferretjans MM, Martín-Cardona C, Lalucat J, Bosch R (2007) Bacterial diversity, composition and dynamics in and around recreational coastal areas. Environ Microbiol 9:1913–1929
Olsen SR (1954) Estimation of available phosphorus in soils by extraction with sodium bicarbonate. US Department of Agriculture
Parada AE, Needham DM, Fuhrman JA (2016) Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. https://doi.org/10.1111/1462-2920.13023
Parks DH, Beiko RG (2010) Identifying biologically relevant differences between metagenomic communities. Bioinform 26:715–721
Peixoto R, Chaer GM, Carmo FL, Araujo FV, Paes JE, Volpon A, Santiago GA, Rosado AS (2011) Bacterial communities reflect the spatial variation in pollutant levels in Brazilian mangrove sediment. Anton Leeuw Int J G 99:341–354. https://doi.org/10.1007/s10482-010-9499-0
Perera K, Amarasinghe M, Somaratna S (2013) Vegetation structure and species distribution of mangroves along a soil salinity gradient in a micro tidal estuary on the north-western coast of Sri Lanka. American J Mar Sci 7–15
Pinhassi J, Sala MM, Havskum H, Peters F, Guadayol O, Malits A, Marrasé C (2004) Changes in bacterioplankton composition under different phytoplankton regimens. Appl Environ Microbiol 70:6753–6766
Qu W, Lin D, Zhang Z, Di W, Gao B, Zeng R (2018) Metagenomics investigation of agarlytic genes and genomes in mangrove sediments in China: a potential repertory for carbohydrate-active enzymes. Front Microbiol 9:1864. https://doi.org/10.3389/fmicb.2018.01864
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Rampadarath S, Bandhoa K, Puchooa D, Jeewon R, Bal S (2018) Metatranscriptomics analysis of mangroves habitats around Mauritius. World J Microbiol Biotechnol 34:59. https://doi.org/10.1007/s11274-018-2442-7
Rasool F, Tunio S, Hasnain S, Ahmad E (2002) Mangrove conservation along the coast of Sonmiani, Balochistan, Pakistan. Trees 16:213–217
Reyes R, Fortes M (1985) Litter production and leaf litter decomposition rates of mangrove trees in Pagbilao, Quezon. Report on microbial aspects of nutrient cycling in mangrove environments, UNDP/UNESCO (RAS/79/002). Philippines, Manila, pp 72–73
Richards L (1954) Diagnosis and improvement of saline and alkali soils (USDA). Handbook. 60. US. Govt., Printing Office, Washington
Rishan ST, Kline RJ, Rahman MS (2023) Applications of environmental DNA (eDNA) to detect subterranean and aquatic invasive species: a critical review on the challenges and limitations of eDNA metabarcoding. Environ Adv 100370
Rivas-Marin E, Canosa I, Devos DP (2016) Evolutionary cell biology of division mode in the bacterial Planctomycetes-Verrucomicrobia- Chlamydiae Superphylum. Front Microbiol 7:1964. https://doi.org/10.3389/fmicb.2016.01964
Rocha LL, Colares GB, Nogueira VL, Paes FA, Melo VM (2016) Distinct habitats select particular bacterial communities in mangrove sediments. Int J Microbiol 2016:3435809. https://doi.org/10.1155/2016/3435809
Roger AJ, Munoz-Gomez SA, Kamikawa R (2017) The origin and diversification of mitochondria. Curr Biol 27:R1177–R1192. https://doi.org/10.1016/j.cub.2017.09.015
Rong X, Zhang W, Xu B, Li P, Wu H, Jin Y, Wang R (2020) Comparative analysis of bacterial community diversity between soil and water of Dongzhai Harbor Mangrove reserve using 16S rRNA gene sequencing and shotgun metagenomic sequencing. Res Sq (Preprint). https://doi.org/10.21203/rs.3.rs-114741/v1
Saifullah S, Rasool F (2002) Mangroves of Miani Hor lagoon on the north Arabian Sea coast of Pakistan. Pak J Bot 34:303–310
Saunders D, Kalff J (2001) Denitrification rates in the sediments of Lake Memphremagog, Canada–USA. Water Res 35:1897–1904. https://doi.org/10.1016/S0043-1354(00)00479-6
Schlaeppi K, Ronchi F, Leib SL, Erb M, Ramette A (2020) Evaluation of primer pairs for microbiome profiling from soils to humans within the One Health framework. Mol Ecol Resour 20:1558–1571
Schloss PD, Handelsman J (2008) A statistical toolbox for metagenomics: assessing functional diversity in microbial communities. BMC Bioinform 9:1–15
Schofield R, Taylor AW (1955) The measurement of soil pH. Soil Sci Soc America J 19:164–167
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genom Biol 12:1–18
Shea MM, Kuppermann J, Rogers MP, Smith DS, Edwards P, Boehm AB (2023) Systematic review of marine environmental DNA metabarcoding studies: toward best practices for data usability and accessibility. PeerJ 11:e14993
Shiau Y-J, Chiu C-Y (2020) Biogeochemical processes of C and N in the soil of mangrove forest ecosystems. Forests 11:492
Simon C, Daniel R (2011) Metagenomic analyses: past and future trends. Appl Environ Microbiol 77:1153–1161. https://doi.org/10.1128/AEM.02345-10
Smith MW, Zeigler Allen L, Allen AE, Herfort L, Simon HM (2013) Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front Microbiol 4:120
Spain AM, Krumholz LR, Elshahed MS (2009) Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J 3:992–1000
Steele HL, Streit WR (2005) Metagenomics: advances in ecology and biotechnology. FEMS Microbiol Lett 247:105–111. https://doi.org/10.1016/j.femsle.2005.05.011
Stewart EJ (2012) Growing unculturable bacteria. J Bacteriol 194:4151–4160. https://doi.org/10.1128/JB.00345-12
Suwa R, Deshar R, Hagihara A (2009) Forest structure of a subtropical mangrove along a river inferred from potential tree height and biomass. Aquat Bot 91:99–104
Thompson CE, Beys-da-Silva WO, Santi L, Berger M, Vainstein MH, Guima Raes JA, Vasconcelos AT (2013) A potential source for cellulolytic enzyme discovery and environmental aspects revealed through metagenomics of Brazilian mangroves. AMB Express 3:65. https://doi.org/10.1186/2191-0855-3-65
Tiwari N, Bansal M, Sharma JG (2021) Metagenomics: a powerful lens viewing the microbial world. In Wastewater Treatment Reactors. Elsevier, pp 309–339
Vazquez P, Holguin G, Puente M, Lopez-Cortes A, Bashan Y (2000) Phosphate-solubilizing microorganisms associated with the rhizosphere of mangroves in a semiarid coastal lagoon. Biol Fertil Soils 30:460–468
Vieira RP, Gonzalez AM, Cardoso AM, Oliveira DN, Albano RM, Clementino MM, Martins OB, Paranhos R (2008) Relationships between bacterial diversity and environmental variables in a tropical marine environment, Rio de Janeiro. Environ Microbiol 10:189–199
Wakley A, Black C (1934) Determination of organic matter in the soil by chromic acid digesion. Soil Sci 63:251–264
Walkley A (1947) A critical examination of a rapid method for determining organic carbon in soils—effect of variations in digestion conditions and of inorganic soil constituents. Soil Sci 63:251–264
Wang S, Hou W, Dong H, Jiang H, Huang L, Wu G, Zhang C, Song Z, Zhang Y, Ren H, Zhang J, Zhang L (2013) Control of temperature on microbial community structure in hot springs of the Tibetan Plateau. PLoS ONE 8:e62901. https://doi.org/10.1371/journal.pone.0062901
Wee AKS, Mori GM, Lira CF, Núñez-Farfán J, Takayama K, Faulks L, Shi S, Tsuda Y, Suyama Y, Yamamoto T, Iwasaki T, Nagano Y, Wang Z, Watanabe S, Kajita T (2019) The integration and application of genomic information in mangrove conservation. Conserv Biol 33:206–209. https://doi.org/10.1111/cobi.13140
Wu P, Xiong X, Xu Z, Lu C, Cheng H, Lyu X, Zhang J, He W, Deng W, Lyu Y, Lou Q, Hong Y, Fang H (2016) Bacterial communities in the rhizospheres of three mangrove tree species from Beilun Estuary, China. PLoS ONE 11:e0164082. https://doi.org/10.1371/journal.pone.0164082
Yan B, Hong K, Yu ZN (2006) Archaeal communities in mangrove soil characterized by 16S rRNA gene clones. J Microbiol 44:566–571
Yang J, Gao J, Cheung A, Liu B, Schwendenmann L, Costello MJ (2013) Vegetation and sediment characteristics in an expanding mangrove forest in New Zealand. Estuar Coast Shelf Sci 134:11–18. https://doi.org/10.1016/j.ecss.2013.09.017
Yeo SK, Huggett MJ, Eiler A, Rappé MS (2013) Coastal bacterioplankton community dynamics in response to a natural disturbance. PLoS ONE 8:e56207
Yuan SY, Chang BV (2007) Anaerobic degradation of five polycyclic aromatic hydrocarbons from river sediment in Taiwan. J Environ Sci Health B 42:63–69
Zhang H, Sekiguchi Y, Hanada S, Hugenholtz P, Kim H, Kamagata Y, Nakamura K (2003) Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int J Syst Evol Microbiol 53:1155–1163. https://doi.org/10.1099/ijs.0.02520-0
Zulfa A, Norizah K, Hamdan O, Faridah-Hanum I, Rhyma P, Fitrianto A (2021) Spectral signature analysis to determine mangrove species delineation structured by anthropogenic effects. Ecol Indic 130:108148
Funding
The work is supported in part under Xinjiang Uygur Autonomous Region, regional coordinated Innovation Project and Shanghai Cooperation Organization Science and Technology Partnership Program China (No. 2021E01018) and Innovation Group Project of Southern Marine Science andEngineering Guangdong Laboratory (Zhuhai) (Nos. 311021004, 311021006). The research was also supported by the Chinese Academy of Sciences President’s International Fellowship Initiative (Grant No.2020VBA0020). WJL was also supported by the Introduction project of high-level talents in Xinjiang Uygur Autonomous Region.
Author information
Authors and Affiliations
Contributions
RB, IA, and BU contributed to conception and design of the study. RB collected samples. ME performed physicochemical analysis of soil samples. RB, AAl, SL, ZL, and JL performed the experiments. RB, SL, JL, and IA organized the database. RB, AAl, SL, ZL, JL, and AAm contributed to the statistical data analysis and finalizing the figures. RB, AAl, AAm, and IA wrote the first draft of the manuscript. AAm, AAl, JL, and YHL wrote sections of the manuscript. IA and WJL managed funds for research and supervised the experiments. All the authors contributed to manuscript revision, read, and approved the submitted version.
Corresponding authors
Ethics declarations
Ethical statement
It is to state that all the authors agreed to the contents of the manuscript and agreed that we (Iftikhar Ahmed and Wen-Jun Li) would act as corresponding authors for the publication. It is also declared that there is no conflict of interest with anyone, and we do not have choice for any editor and /or reviewer to be included or excluded for reviewing this manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bushra, R., Ahmed, I., Li, JL. et al. Untapped rich microbiota of mangroves of Pakistan: diversity and community compositions. Folia Microbiol (2023). https://doi.org/10.1007/s12223-023-01095-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12223-023-01095-3