Abstract
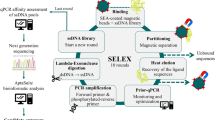
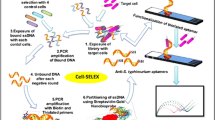
Salmonella enterica subsp. enterica ser. enteritidis and Salmonella enterica subsp. enterica ser. typhimurium are the most common and severe food-borne pathogens responsible for causing salmonellosis in humans and animals. The development of an early and ultra-sensitive detection system is the first critical step in controlling this disease. To accomplish this, we used the cell systematic evolution of ligands by exponential enrichment (Cell-SELEX) technique to identify single-stranded DNA (ssDNA) aptamers to be used as detection probes that can specifically bind to S. enteritidis and S. typhimurium. A total of 12 target-specific ssDNA aptamers were obtained through ten rounds of Cell-SELEX under stringent selection conditions, and negative selection further enhanced the selectivity among these aptamers. Aptamer specificity was investigated using the gram-negative bacteria E. coli and P. aeruginosa and was found to be much higher towards S. enteritidis and S. typhimurium. Importantly, three candidate aptamers demonstrated higher binding affinities and the dissociation constants (Kd) were found to be in the range of nanomolar to submicromolar levels. Furthermore, individual aptamers were conjugated onto polyvalent directed aptamer polymer, which led to 100-fold increase in binding affinity compared to the individual aptamers alone. Taken together, this study reports the identification of higher affinity and specificity ssDNA aptamers (30mer), which may be useful as capture and detection probes in biosensor-based detection systems for salmonellosis.





Similar content being viewed by others
References
Pui, C. F., Wong, W. C., Chai, L. C., Tunung, R., Jeyaletchumi, P. N., Hidayah, M. S., Ubong, A., Farinazleen, M. G., Cheah, Y. K., & Son, R. (2011). Salmonella: a foodborne pathogen. International Food Research Journal, 18, 465–473.
Pegues, D. A., Ohl, M. E., & Miller, S. I. (2005). Salmonella species, including Salmonella typhi. In G. L. Mandell, J. E. Bennet, & R. Dolin (Eds.), Principles and practice of infectious diseases (6th ed., pp. 2636–2654). New York: Churchill Livingstone, Elsevier.
Helms, M., Ethelberg, S., & Molbak, K. (2005). International Salmonella typhimurium DT104 infections, 1992–2001. Emerging Infectious Diseases, 11, 859–867.
Dechet, A. M., Scalla, E., Gensheimer, K., Hoekstra, R., Gunderman-King, Lockett, J. J., Wrigley, D., Chege, W., & Sobel, J. (2006). Multistate Working Group. Outbreak of multidrug-resistant Salmonella enterica serotype Typhimurium Definitive Type 104 infection linked to commercial ground beef, northeastern United States, 2003–2004. Clinical Infectious Diseases, 42, 747–752.
WHO Global Salm-Surv Progress Report II. 2000–2005. WHO Press, World Health Organization, Geneva, Switzerland.
http://www.cdc.gov/nczved/divisions/dfbmd/diseases/salmonellosis/ (last accessed on 23 January 2014).
Lee, D. Y., Lee, E., Min, J. E., Kim, S. H., Oh, H. B., & Park, M. S. (2011). Epidemic by Salmonella I 4,[5],12:i:- and characteristics of isolates in Korea. Infect Chemother, 43, 186–190.
Brenner, F. W., Villar, R. G., Angulo, F. J., Tauxe, & Swaminathan, R. B. (2000). Salmonella nomenclature. Journal of Clinical Microbiology, 38, 2465–2467.
Dziadkowiec, D., Mansfield, L. P., & Forsythe, S. J. (1995). The detection of Salmonella in skimmed milk powder enrichments using conventional methods and immunomagnetic separation. Letters in Applied Microbiology, 20, 361–364.
Techathuvanan, C., Draughon, F. A., & D’Souza, D. H. (2010). Real-time reverse transcriptase PCR for the rapid and sensitive detection of Salmonella typhimurium from pork. Journal of Food Protection, 73, 507–514.
Patel, J. R., Bhagwat, A. A., Sanglay, G. C., & Solomon, M. B. (2006). Rapid detection of Salmonella from hydrodynamic pressure-treated poultry using molecular beacon real-time PCR. Food Microbiology, 23, 39–46.
Oracz, G., Feleszko, W., Golicka, D., Maksymiuk, J., Klonowska, A., & Szajewska, H. (2003). Rapid diagnosis of acute Salmonella gastrointestinal infection. Clinical Infectious Diseases, 36, 112–115.
Holt, P. S., Gast, R. K., & Greene, C. R. (1995). Rapid detection of S. enteritidis in pooled liquid egg samples using a magnetic bead-ELISA system. Journal of Food Protection, 58, 967–972.
Kumar, S., Balakrishna, K., & Batra, H. V. (2008). Enrichment-elisa for detection of Salmonella typhi from food and water samples. Biomedical and Environmental Sciences, 21, 137–143.
Lan, Y. B., Wang, S. Z., Yin, Y. G., Clint Hoffmann, W., & Zheng, X. Z. (2008). Using a surface Plasmon resonance biosensor for rapid detection of Salmonella typhimurium in chicken carcass. Journal of Bionic Engineering, 5, 239–246.
Salam, F., & Tothill, I. E. (2009). Detection of Salmonella typhimurium using an electrochemical immunosensor. Biosensors and Bioelectronics, 24, 2630–2636.
Yuan, J., Tao, Z., Yu, Y., Ma, X., Xia, Y., Wang, L., & Wang, Z. (2014). A visual detection method for Salmonella typhimurium based on aptamer recognition and nanogold labeling. Food Control, 37, 188–192.
Zuo, P., Li, X., Dominguez, D. C., & Ye, B. C. (2013). A PDMS/paper/glass hybrid microfluidic biochipintegrated with aptamer-functionalized graphene oxidenano-biosensors for one-step multiplexed pathogen detection. Lab on a Chip, 13, 3921–3928.
Tombelli, S., Minunni, M., & Mascini, M. (2005). Analytical applications of aptamers. Biosensors and Bioelectronics, 20, 2424–2434.
Irvine, D., Tuerk, C., & Gold, L. (1991). SELEXION. Systematic evolution of nucleic acids by exponential enrichment with integrated optimization by non-linear analysis. Journal of Molecular Biology, 222, 739–761.
Jayasena, S. D. (1999). Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clinical Chemistry, 45, 1628–1650.
Cibiel, A., Dupont, D. M., & Ducongé, F. (2011). Methods to identify aptamers against cell surface biomarkers. Pharmaceuticals, 4, 1216–1235.
Stoltenburg, R., Reinemann, C., & Strehlitz, B. (2007). SELEX—a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomolecular Engineering, 24, 381–403.
Tan, W., Donovan, M. J., & Jiang, J. (2013). Aptamers from cell-based selection for bioanalytical applications. Chemistry Review, 113, 2842–2862.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press.
Zuker, M. (1989). On finding all suboptimal foldings of an RNA molecule. Science, 244, 48–52.
Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research, 31, 3406–3415.
Park, H. Y., Gedi, V. K., Kim, J., Park, H. C., Han, S. N., & Yoon, M. Y. (2011). Ultrasensitive diagnosis for an anthrax-protective antigen based on a polyvalent directed peptide polymer coupled to zinc oxide nanorods. Advanced Materials, 23, 5425–5429.
Chen, F., Zhou, J., Luo, F., Mohammed, A. B., & Zhang, X. L. (2007). Aptamer from whole-bacterium SELEX as new therapeutic reagent against virulent Mycobacterium tuberculosis. Biochemical and Biophysical Research Communications, 357, 743–748.
Lee, Y. J., Han, S. R., Maeng, J. S., Cho, Y. J., & Lee, S. W. (2012). In vitro selection of Escherichia coli O157:H7-specific RNA aptamer. Biochemical and Biophysical Research Communications, 417, 414–420.
Dwivedi, H. P., Derike, S. R., & Jaykus, L. A. (2010). Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Applied Microbiology and Biotechnology, 87, 2323–2334.
Joshi, R., Janagama, H., Dwivedi, H. P., Senthil Kumar, T. M. A., Jaykus, L. A., Schefers, J., & Sreevatsan, S. (2009). Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars. Molecular and Cellular Probes, 23, 20–28.
Moon, J., Kim, G., Lee, S., & Park, S. (2013). Identification of salmonella typhimurium-specific DNA aptamers developed using whole-cell SELEX and FACS analysis. Journal of Microbiological Methods, 95, 162–166.
Ye, M., Hu, J., Peng, M., Liu, J., Liu, J., Liu, H., Zhao, X., & Tan, W. (2012). Generating aptamers by cell-SELEX for applications in molecular medicine. International Journal of Molecular Sciences, 13, 3341–3353.
Kissel, J. D., Held, D. M., Hardy, R. W., & Burke, D. H. (2007). Active site binding and sequence requirements for inhibition of HIV-1 reverse transcriptase by the RT1 family of single-stranded DNA aptamers. Nucleic Acids Research, 35, 5039–5050.
Fischer, N. O., Tok, J. B. H., & Tarasow, T. M. (2008). Massively parallel interrogation of aptamer sequence, structure, and function. PloS One, 3, e2720. doi:10.1371/journal.pone.0002720.
Vandenengel, J. E., & Morse, D. P. (2009). Mutational analysis of a signaling aptamer suggests a mechanism for ligand-triggered structure-switching. Biochemical and Biophysical Research Communications, 378, 51–56.
Acknowledgments
This work was supported by the “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ907052)” funded by the Rural Development Administration and by the “Basic Science Research Program through the National Research Foundation of Korea (NRF)” funded by the Ministry of Education, Science and Technology (2012R1A1A2008516), Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Highlights
Identified ssDNA aptamers that selectively bind to S. enteritidis and S. typhimurium.
The candidate aptamers showed high affinity in the nanomolar to sub micromolar range.
Development of conjugated aptamers (PDAP) showed enhanced affinity and specificity.
The identified short 30-mer DNA aptamers can be useful as sensitive detection probes.
Hae-Chul Park and Irshad Ahmed Baig has contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Scheme of PDAP synthesis. a Schematic representation of PDAP (polyvalent directed aptamer polymer). b Schematic representation of PDAP construction. (1) Aptamers were double modified with 5′-FAM and 3′-SH, where FAM acts as the signal probe and the SH group is used as the anchor molecule between the aptamer and SPDP by disulfide bond formation. SPDP is reactive toward amines through the succinimide group and toward sulfhydryl groups through the pyridylthiol group. (2) SPDP-mediated activation of PDL was done by mixing the SPDP-PDL solution at room temperature. The complex was harvested using a gel filtration column. (3) 5′-FAM and 3′-SH labeled aptamers were added to SPDP activated PDL solution. (DOCX 174 kb)
Rights and permissions
About this article
Cite this article
Park, HC., Baig, I.A., Lee, SC. et al. Development of ssDNA Aptamers for the Sensitive Detection of Salmonella typhimurium and Salmonella enteritidis . Appl Biochem Biotechnol 174, 793–802 (2014). https://doi.org/10.1007/s12010-014-1103-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-1103-z