Abstract
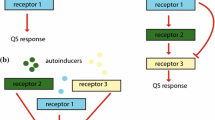
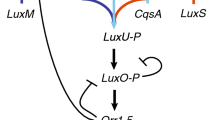
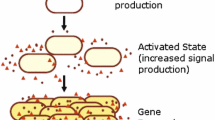
Many bacteria alter their behaviors as a function of population density, via a process known as quorum sensing (QS). QS is achieved by the synthesis and detection of diffusible signal molecules, often involving complex signal transduction pathways and regulatory networks. Mathematical models have been developed to investigate a number of aspects of QS, resulting in a wide range of model structures; many have focused on either the molecular or the population scale. In this paper, I show that many published models fail to satisfy physical constraints (such as conservation of matter) or rely on a priori assumptions that may not be valid. I present new, simple models of canonical Gram-negative and Gram-positive QS systems, in both well-mixed and biofilm populations, focusing on the interaction between molecular and population processes. I show that this interaction may be crucial for several important features of QS, including bistability and the localization of QS in space. The results highlight the need to link molecular and population processes carefully in QS models, provide a general framework for understanding the behavior of complex system-specific models, and suggest new directions for both theoretical and experimental work.







Similar content being viewed by others
References
Anetzberger, C., Pirch, T., & Jung, K. (2009). Heterogeneity in quorum sensing-regulated bioluminescence of Vibrio harveyi. Mol. Microbiol., 73(2), 267–277.
Anguige, K., King, J. R., Ward, J. P., & Williams, P. (2004). Mathematical modelling of therapies targeted at bacterial quorum sensing. Math. Biosci., 192(1), 39–83.
Anguige, K., King, J. R., & Ward, J. P. (2005). Modelling antibiotic- and anti-quorum sensing treatment of a spatially-structured Pseudomonas aeruginosa population. J. Math. Biol., 51(5), 557–594.
Anguige, K., King, J. R., & Ward, J. P. (2006). A multi-phase mathematical model of quorum sensing in a maturing Pseudomonas aeruginosa biofilm. Math. Biosci., 203(2), 240–276.
Balagadde, F. K., Song, H., Ozaki, J., Collins, C. H., Barnet, M., Arnold, F. H., Quake, S. R., & You, L. (2008). A synthetic Escherichia coli predator-prey ecosystem. Mol. Syst. Biol., 4, 187.
Banik, S. K., Fenley, A. T., & Kulkarni, R. V. (2009). A model for signal transduction during quorum sensing in Vibrio harveyi. Phys. Biol., 6(4), 046008.
Bassler, B. L. (1999). How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol., 2(6), 582–587.
Bedford, A., & Drumheller, D. S. (1983). Theory of immiscible and structured mixtures. Int. J. Eng. Sci., 21, 863–960.
Bejerano-Sagie, M., & Xavier, K. B. (2007). The role of small RNAs in quorum sensing. Curr. Opin. Microbiol., 10(2), 189–198.
Brown, D. (2010). A mathematical model of the Gac/Rsm quorum sensing network in Pseudomonas fluorescens. Biosystems, 101(3), 200–212.
Capaldi, F. M. (2012). Continuum mechanics: constitutive modeling of structural and biological materials.
Chopp, D. L., Kirisits, M. J., Moran, B., & Parsek, M. R. (2002). A mathematical model of quorum sensing in a growing bacterial biofilm. J. Ind. Microbiol. Biotech., 29(6), 339–346.
Chopp, D. L., Kirisits, M. J., Moran, B., & Parsek, M. R. (2003). The dependence of quorum sensing on the depth of a growing biofilm. Bull. Math. Biol., 65(6), 1053–1079.
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R., & Lappin-Scott, H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol., 49, 711–745.
Cox, C. D., Peterson, G. D., Allen, M. S., Lancaster, J. M., McCollum, J. M., Austin, D., Yan, L., Sayler, G. S., & Simpson, M. L. (2003). Analysis of noise in quorum sensing. Omics. J. Integr. Biol., 7(3), 317–334.
Darch, S. E., West, S. A., Winzer, K., & Diggle, S. P. (2012). Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc. Natl. Acad. Sci. USA, 109(21), 8259–8263.
Dockery, J. D., & Keener, J. P. (2001). A mathematical model for quorum sensing in Pseudomonas aeruginosa. Bull. Math. Biol., 63(1), 95–116.
Fagerlind, M. G., Rice, S. A., Nilsson, P., Harlen, M., James, S., Charlton, T., & Kjelleberg, S. (2003). The role of regulators in the expression of quorum-sensing signals in Pseudomonas aeruginosa. J. Mol. Microbiol. Biotechnol., 6(2), 88–100.
Fagerlind, M. G., Nilsson, P., Harlen, M., Karlsson, S., Rice, S. A., & Kjelleberg, S. (2005). Modeling the effect of acylated homoserine lactone antagonists in Pseudomonas aeruginosa. Biosystems, 80(2), 201–213.
Fenley, A. T., Banik, S. K., & Kulkarni, R. V. (2011). Computational modeling of differences in the quorum sensing induced luminescence phenotypes of Vibrio harveyi and Vibrio cholerae. J. Theor. Biol., 274(1), 145–153.
Fuqua, C., Parsek, M. R., & Greenberg, E. P. (2001). Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet., 35, 439–468.
Garcia-Ojalvo, J., Elowitz, M. B., & Strogatz, S. H. (2004). Modeling a synthetic multicellular clock: repressilators coupled by quorum sensing. Proc. Natl. Acad. Sci. USA, 101(30), 10955–10960.
Gonzalez Barrios, A. F., Covo, V., Medina, L. M., Vives-Florez, M., & Achenie, L. (2009). Quorum quenching analysis in Pseudomonas aeruginos and Escherichia coli: network topology and inhibition mechanism effect on the optimized inhibitor dose. Bioprocess. Biosyst. Eng., 32, 545–556.
Goryachev, A. B. (2011). Understanding bacterial cell-cell communication with computational modeling. Chem. Rev., 111(1), 238–250.
Goryachev, A. B., Toh, D. J., Wee, K. B., Lee, T., Zhang, H. B., & Zhang, L. H. (2005). Transition to quorum sensing in an agrobacterium population: a stochastic model. PLoS Comput. Biol., 1(4), e37.
Goryachev, A. B., Toh, D. J., & Lee, T. (2006). Systems analysis of a quorum sensing network: design constraints imposed by the functional requirements, network topology and kinetic constants. Biosystems, 83(2–3), 178–187.
Gustafsson, E., Nilsson, P., Karlsson, S., & Arvidson, S. (2004). Characterizing the dynamics of the quorum-sensing system in Staphylococcus aureus. J. Mol. Microbiol. Biotechnol., 8(4), 232–242.
Hasty, J., Pradines, J., Dolnik, M., & Collins, J. J. (2000). Noise-based switches and amplifiers for gene expression. Proc. Natl. Acad. Sci. USA, 97(5), 2075–2080.
Heeb, S., & Haas, D. (2001). Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact., 14(12), 1351–1363.
Hense, B. A., Kuttler, C., Muller, J., Rothballer, M., Hartmann, A., & Kreft, J. U. (2007). Does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol., 5(3), 230–239.
Hong, D., Saidel, W. M., Man, S., & Martin, J. V. (2007). Extracellular noise-induced stochastic synchronization in heterogeneous quorum sensing network. J. Theor. Biol., 245(4), 726–736.
Jabbari, S., King, J. R., Koerber, A. J., & Williams, P. (2010a). Mathematical modelling of the agr operon in Staphylococcus aureus. J. Math. Biol., 61(1), 17–54.
Jabbari, S., King, J. R., & Williams, P. (2010b). A mathematical investigation of the effects of inhibitor therapy on three putative phosphorylation cascades governing the two-component system of the agr operon. Math. Biosci., 225(2), 115–131.
Jabbari, S., Heap, J. T., & King, J. R. (2011). Mathematical modelling of the sporulation-initiation network in Bacillus subtilis revealing the dual role of the putative quorum-sensing signal molecule PhrA. Bull. Math. Biol., 73(1), 181–211.
Jabbari, S., King, J. R., & Williams, P. (2012a). Cross-strain quorum sensing inhibition by Staphylococcus aureus. Part 1: a spatially homogeneous model. Bull. Math. Biol., 74(6), 1292–1325.
Jabbari, S., King, J. R., & Williams, P. (2012b). Cross-strain quorum sensing inhibition by Staphylococcus aureus. Part 2: a spatially inhomogeneous model. Bull. Math. Biol., 74(6), 1326–1353.
James, S., Nilsson, P., James, G., Kjelleberg, S., & Fagerstrom, T. (2000). Luminescence control in the marine bacterium Vibrio fischeri: an analysis of the dynamics of lux regulation. J. Mol. Biol., 296(4), 1127–1137.
Karlsson, D., Karlsson, S., Gustafsson, E., Normark, B. H., & Nilsson, P. (2007). Modeling the regulation of the competence-evoking quorum sensing network in Streptococcus pneumoniae. Biosystems, 90(1), 211–223.
Kim, J. R., & Cho, K. H. (2006). The multi-step phosphorelay mechanism of unorthodox two-component systems in E. coli realizes ultrasensitivity to stimuli while maintaining robustness to noises. Comput. Biol. Chem., 30(6), 438–444.
Koerber, A. J., King, J. R., Ward, J. P., Williams, P., Croft, J. M., & Sockett, R. E. (2002). A mathematical model of partial-thickness burn-wound infection by Pseudomonas aeruginosa: quorum sensing and the build-up to invasion. Bull. Math. Biol., 64(2), 239–259.
Koerber, A. J., King, J. R., & Williams, P. (2005). Deterministic and stochastic modelling of endosome escape by Staphylococcus aureus: “quorum” sensing by a single bacterium. J. Math. Biol., 50(4), 440–488.
Kuttler, C., & Hense, B. A. (2008). Interplay of two quorum sensing regulation systems of Vibrio fischeri. J. Theor. Biol., 251(1), 167–180.
Lapouge, K., Schubert, M., Allain, F. H., & Haas, D. (2008). Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol., 67(2), 241–253.
Lenz, A. P., Williamson, K. S., Pitts, B., Stewart, P. S., & Franklin, M. J. (2008). Localized gene expression in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol., 74(14), 4463–4471.
Levsky, J. M., & Singer, R. H. (2003). Gene expression and the myth of the average cell. Trends Cell Biol., 13(1), 4–6.
Mehra, S., Charaniya, S., Takano, E., & Hu, W. S. (2008). A bistable gene switch for antibiotic biosynthesis: the butyrolactone regulon in Streptomyces coelicolor. PLoS ONE, 3(7), e2724.
Mehta, P., Goyal, S., Long, T., Bassler, B. L., & Wingreen, N. S. (2009). Information processing and signal integration in bacterial quorum sensing. Mol. Syst. Biol., 5, 325.
Melke, P., Sahlin, P., Levchenko, A., & Jonsson, H. (2010). A cell-based model for quorum sensing in heterogeneous bacterial colonies. PLoS Comput. Biol., 6(6), e1000819.
Muller, J., Kuttler, C., Hense, B. A., Rothballer, M., & Hartmann, A. (2006). Cell-cell communication by quorum sensing and dimension-reduction. J. Math. Biol., 53(4), 672–702.
Muller, J., Kuttler, C., & Hense, B. A. (2008). Sensitivity of the quorum sensing system is achieved by low pass filtering. Biosystems, 92(1), 76–81.
Ng, W. L., & Bassler, B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet., 43, 197–222.
Nilsson, P., Olofsson, A., Fagerlind, M., Fagerstrom, T., Rice, S., Kjelleberg, S., & Steinberg, P. (2001). Kinetics of the AHL regulatory system in a model biofilm system: how many bacteria constitute a “quorum”? J. Mol. Biol., 309(3), 631–640.
Pai, A., & You, L. (2009). Optimal tuning of bacterial sensing potential. Mol. Syst. Biol., 5, 286.
Pearson, J. P., Van Delden, C., & Iglewski, B. H. (1999). Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol., 181(4), 1203–1210.
Redfield, R. J. (2002). Is quorum sensing a side effect of diffusion sensing? Trends Microbiol., 10(8), 365–370.
Tanouchi, Y., Tu, D., Kim, J., & You, L. (2008). Noise reduction by diffusional dissipation in a minimal quorum sensing motif. PLoS Comput. Biol., 4(8), e1000167.
Viretta, A. U., & Fussenegger, M. (2004). Modeling the quorum sensing regulatory network of human-pathogenic Pseudomonas aeruginosa. Biotechnol. Prog., 20(3), 670–678.
Ward, J. P., King, J. R., Koerber, A. J., Williams, P., Croft, J. M., & Sockett, R. E. (2001). Mathematical modelling of quorum sensing in bacteria. IMA J. Math. Appl. Med. Biol., 18(3), 263–292.
Ward, J. P., King, J. R., Koerber, A. J., Croft, J. M., Sockett, R. E., & Williams, P. (2003). Early development and quorum sensing in bacterial biofilms. J. Math. Biol., 47(1), 23–55.
Ward, J. P., King, J. R., Koerber, A. J., Croft, J. M., Sockett, R. E., & Williams, P. (2004). Cell-signalling repression in bacterial quorum sensing. Math. Med. Biol., 21(3), 169–204.
Williams, J. W., Cui, X., Levchenko, A., & Stevens, A. M. (2008). Robust and sensitive control of a quorum-sensing circuit by two interlocked feedback loops. Mol. Syst. Biol., 4, 234.
Yarwood, J. M., Bartels, D. J., Volper, E. M., & Greenberg, E. P. (2004). Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol., 186(6), 1838–1850.
Xu, K. D., Stewart, P. S., Xia, F., Huang, C. T., & McFeters, G. A. (1998). Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol., 64(10), 4035–4039.
Acknowledgements
I thank Phoebe Lostroh for many interesting conversations about bacterial genetics and quorum sensing. I thank Sara Jabbari for discussing her modeling work with me.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brown, D. Linking Molecular and Population Processes in Mathematical Models of Quorum Sensing. Bull Math Biol 75, 1813–1839 (2013). https://doi.org/10.1007/s11538-013-9870-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-013-9870-1