Abstract
Introduction
The measurement of specific volatile organic compounds in breath has been proposed as a potential diagnostic for a variety of diseases. The most well-studied bacterial lung infection in the breath field is that caused by Pseudomonas aeruginosa.
Objectives
To determine a discriminatory core of molecules in the “breath-print” of mice during a lung infection with four strains of P. aeruginosa (PAO1, PA14, PAK, PA7). Furthermore, we attempted to extrapolate a strain-specific “breath-print” signature to investigate the possibility of recapitulating the genetic phylogenetic groups (Stewart et al. Pathog Dis 71(1), 20–25, 2014. https://doi.org/10.1111/2049-632X.12107).
Methods
Breath was collected into a Tedlar bag and shortly after drawn into a thermal desorption tube. The latter was then analyzed into a comprehensive multidimensional gas chromatography coupled with a time-of-flight mass spectrometer. Random forest algorithm was used for selecting the most discriminatory features and creating a prediction model.
Results
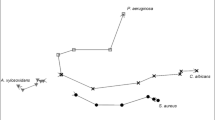
Three hundred and one molecules were significantly different between animals infected with P. aeruginosa, and those given a sham infection (PBS) or inoculated with UV-killed P. aeruginosa. Of those, nine metabolites could be used to discriminate between the three groups with an accuracy of 81%. Hierarchical clustering showed that the signature from breath was due to a specific response to live bacteria instead of a generic infection response. Furthermore, we identified ten additional volatile metabolites that could differentiate mice infected with different strains of P. aeruginosa. A phylogram generated from the ten metabolites showed that PAO1 and PA7 were the most distinct group, while PAK and PA14 were interspersed between the former two groups.
Conclusions
To the best of our knowledge, this is the first study to report on a ‘core’ murine breath print, as well as, strain level differences between the compounds in breath. We provide identifications (by running commercially available analytical standards) to five breath compounds that are predictive of P. aeruginosa infection.





Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Bean, H. D., Jiménez-Díaz, J., Zhu, J., & Hill, J. E. (2015). Breathprints of model murine bacterial lung infections are linked with immune response. European Respiratory Journal, 45(1), 181–190. https://doi.org/10.1183/09031936.00015814.
Bean, H. D., Rees, C. A., & Hill, J. E. (2016). Comparative analysis of the volatile metabolomes of Pseudomonas aeruginosa clinical isolates. Journal of Breath Research, 10, 047102. https://doi.org/10.1088/1752-7155/10/4/047102.
Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological), 57(1), 289–300.
Breiman, L. (2001). Random forests. Machine Learning, 45(1), 5–32. https://doi.org/10.1023/A:1010933404324.
Franchina, F. A., Mellors, T. R., Aliyeva, M., Wagner, J., Daphtary, N., Lundblad, L. K. A., et al. (2018). Towards the use of breath for detecting mycobacterial infection: A case study in a murine model. Journal of Breath Research, 12(2), 26008. https://doi.org/10.1088/1752-7163/aaa016.
Frimmersdorf, E., Horatzek, S., Pelnikevich, A., Wiehlmann, L., & Schomburg, D. (2010). How Pseudomonas aeruginosa adapts to various environments: A metabolomic approach. Environmental Microbiology, 12(6), 1734–1747. https://doi.org/10.1111/j.1462-2920.2010.02253.x.
Giorgio, A., De Stradis, A., Lo Cantore, P., & Iacobellis, N. S. (2015). Biocide effects of volatile organic compounds produced by potential biocontrol rhizobacteria on Sclerotinia sclerotiorum. Frontiers in Microbiology, 6, 1056. https://doi.org/10.3389/fmicb.2015.01056.
Gisbert, J. P., & Pajares, J. M. (2004). Review article: 13C-urea breath test in the diagnosis of Helicobacter pylori infection—A critical review. Alimentary Pharmacology and Therapeutics, 20(10), 1001–1017. https://doi.org/10.1111/j.1365-2036.2004.02203.x.
Goeminne, P. C., Vandendriessche, T., Van Eldere, J., Nicolai, B. M., Hertog, M. L., A., T., M., & Dupont, L. J. (2012). Detection of Pseudomonas aeruginosa in sputum headspace through volatile organic compound analysis. Respiratory Research, 13, 87. https://doi.org/10.1186/1465-9921-13-87.
Haick, H., Broza, Y. Y., Mochalski, P., Ruzsanyi, V., & Amann, A. (2014). Assessment, origin, and implementation of breath volatile cancer markers. Chemical Society Reviews, 43, 1423–1449. https://doi.org/10.1039/c3cs60329f.
Jacobs, M. A., Alwood, A., Thaipisuttikul, I., Spencer, D., Haugen, E., Ernst, S., et al. (2003). Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America, 100(24), 14339–14344. https://doi.org/10.1073/pnas.2036282100.
Kharitonov, S. A., Yates, D., Robbins, R. A., Logan-Sinclair, R., Shinebourne, E. A., & Barnes, P. J. (1994). Increased nitric oxide in exhaled air of asthmatic patients. Lancet, 343(8890), 133–135.
Kruskal, W. H., & Wallis, W. A. (1952). Use of ranks in one-criterion variance analysis. Journal of the American Statistical Association, 47(260), 583–621.
Lyczak, J. B., Cannon, C. L., & Pier, G. B. (2000). Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes and Infection, 2(9), 1051–1060. https://doi.org/10.1016/S1286-4579(00)01259-4.
Minamishima, Y., Takeya, K., Ohnishi, Y., & Amako, K. (1968). Physicochemical and biological properties of fibrous Pseudomonas bacteriophages. Journal of Virology, 2(3), 208–213.
Nasir, M., Bean, H. D., Smolinska, A., Rees, C. A., Zemanick, E. T., & Hill, J. E. (2018). Volatile molecules from bronchoalveolar lavage fluid can ‘rule-in’ Pseudomonas aeruginosa and ‘rule-out’ Staphylococcus aureus infections in cystic fibrosis patients. Scientific Reports, 8(1), 826. https://doi.org/10.1038/S41598-017-18491-8.
Patti, G. J., Yanes, O., & Siuzdak, G. (2012). Metabolomics: The apogee of the omics trilogy. Nature Reviews Molecular Cell Biology, 13(4), 263–269. https://doi.org/10.1038/nrm3314.
Phillips, M., Boehmer, J. P., Cataneo, R. N., Cheema, T., Greenberg, J., Kobashigawa, J., et al. (2004). Heart allograft rejection: Detection with breath alkanes in low levels (the HARDBALL Study) methods: Results. The Journal of heart and lung Transplantation, 23(6), 701–708. https://doi.org/10.1016/j.healun.2003.07.017.
Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G., & Ausubel, F. M. (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science, 268(5219), 1899–1902.
Robroeks, C. M. H. H. T., van Berkel, J. J. B. N., Dallinga, J. W., Jöbsis, Q., Zimmermann, L. J. I., Hendriks, H. J. E., et al. (2010). Metabolomics of volatile organic compounds in cystic fibrosis patients and controls. Pediatric Research, 68(1), 75–80. https://doi.org/10.1203/PDR.0b013e3181df4ea0.
Roy, P. H., Tetu, S. G., Larouche, A., Elbourne, L., Tremblay, S., Ren, Q., et al. (2010). Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PloS ONE, 5(1), e8842. https://doi.org/10.1371/journal.pone.0008842.
Sethi, S., Nanda, R., & Chakraborty, T. (2013). Clinical application of volatile organic compound analysis for detecting infectious diseases. Clinical Microbiology Reviews, 26(3), 462–475. https://doi.org/10.1128/CMR.00020-13.
Shestivska, V., Spanel, P., Dryahina, K., Sovova, K., Smith, D., Musilek, M., & Nemec, A. (2012). Variability in the concentrations of volatile metabolites emitted by genotypically different strains of Pseudomonas aeruginosa. Journal of Applied Microbiology, 113, 701–713. https://doi.org/10.1111/j.1365-2672.2012.05370.x.
Silkoff, P. E., Carlson, M., Bourke, T., Katial, R., Ögren, E., & Szefler, S. J. (2004). The Aerocrine exhaled nitric oxide monitoring system NIOX is cleared by the US Food and Drug Administration for monitoring therapy in asthma. The Journal of Allergy and Clinical Immunology, 114(5), 1241–1256. https://doi.org/10.1016/j.jaci.2004.08.042.
Smolinska, A., Hauschild, A.-C., Fijten, R. R. R., Dallinga, J. W., Baumbach, J., & van Schooten, F. J. (2014). Current breathomics—A review on data pre-processing techniques and machine learning in metabolomics breath analysis. Journal of Breath Research, 8(2), 027105. https://doi.org/10.1088/1752-7155/8/2/027105.
Stewart, L., Ford, A., Sangal, V., Jeukens, J., Boyle, B., Kukavica-Ibrulj, I., et al. (2014). Draft genomes of 12 host-adapted and environmental isolates of Pseudomonas aeruginosa and their positions in the core genome phylogeny. Pathogens and Disease, 71(1), 20–25. https://doi.org/10.1111/2049-632X.12107.
Tranchida, P. Q., Maimone, M., Purcaro, G., Dugo, P., & Mondello, L. (2015). The penetration of green sample-preparation techniques in comprehensive two-dimensional gas chromatography. TrAC Trends in Analytical Chemistry, 71, 74–84. https://doi.org/10.1016/j.trac.2015.03.011.
Zhu, J., Bean, H. D., Jimenez-Diaz, J., & Hill, J. E. (2013a). Secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting of multiple bacterial lung pathogens, a mouse model study. Journal of Applied Physiology, 114(11), 1544–1549. https://doi.org/10.1152/japplphysiol.00099.2013.
Zhu, J., Bean, H. D., Wargo, M. J., Leclair, L. W., & Hill, J. E. (2014). Detecting bacterial lung infections: In vivo evaluation of in vitro volatile fingerprints. Journal of Breath Research, 7(1), 016003. https://doi.org/10.1088/1752-7155/7/1/016003.Detecting.
Zhu, J., Jiménez-Díaz, J., Bean, H. D., Daphtary, N. A., Aliyeva, M. I., Lundblad, L. K. A., & Hill, J. E. (2013b). Robust detection of P. aeruginosa and S. aureus acute lung infections by secondary electrospray ionization-mass spectrometry (SESI-MS) breathprinting: From initial infection to clearance. Journal of Breath Research, 7(3), 037106. https://doi.org/10.1088/1752-7155/7/3/037106.
Acknowledgements
Financial support for this work was provided by Hitchcock Foundation and the National Institutes of Health (NIH, Project # R21AI12107601). MN and CAR were supported by the Burroughs Wellcome Fund institutional program grant unifying population and laboratory based sciences to Dartmouth College (Grant#1014106). CAR was additionally supported by a T32 training grant (T32LM012204, PI: Tor D Tosteson).
Author information
Authors and Affiliations
Contributions
Conception and design: GP, JEH, LKAL, MJW; Acquisition of the data: GP, MN, FAF, CR, AM, DN; Analysis and interpretation of the data: MN, GP, FAF; Writing and review of the manuscript: GP, MN, FAF, CR, MJW, LKAL, JEH.
Corresponding author
Ethics declarations
Conflict of interest
All authors report no potential conflicts of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Research involving animal rights
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Purcaro, G., Nasir, M., Franchina, F.A. et al. Breath metabolome of mice infected with Pseudomonas aeruginosa. Metabolomics 15, 10 (2019). https://doi.org/10.1007/s11306-018-1461-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-018-1461-6