Abstract
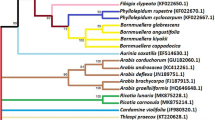
Oryzoideae (Poaceae) plants have economic and ecological value. However, the phylogenetic position of some plants is not clear, such as Hygroryza aristata (Retz.) Nees. and Porteresia coarctata (Roxb.) Tateoka (syn. Oryza coarctata). Comprehensive molecular phylogenetic studies have been carried out on many genera in the Poaceae. The different DNA sequences, including nuclear and chloroplast sequences, had been extensively employed to determine relationships at both higher and lower taxonomic levels in the Poaceae. Chloroplast DNA ndhF gene and atpB-rbcL spacer were used to construct phylogenetic trees and estimate the divergence time of Oryzoideae, Bambusoideae, Panicoideae, Pooideae and so on. Complete sequences of atpB-rbcL and ndhF were generated for 17 species representing six species of the Oryzoideae and related subfamilies. Nicotiana tabacum L. was the outgroup species. The two DNA datasets were analyzed, using Maximum Parsimony and Bayesian analysis methods. The molecular phylogeny revealed that H. aristata (Retz.) Nees was the sister to Chikusichloa aquatica Koidz. Moreover, P. coarctata (Roxb.) Tateoka was in the genus Oryza. Furthermore, the result of evolution analysis, which based on the ndhF marker, indicated that the time of origin of Oryzoideae might be 31 million years ago.




Similar content being viewed by others
References
Gaut B (2002) Evolutionary dynamics of grass genomes. New Phytol 154:15–28
Shantz H (1954) The place of grasslands in the earth’s cover. Ecology 35:143–145
Aggarwal R, Brar D, Nandi S et al (1999) Phylogenetic relationships among Oryza species revealed by AFLP markers. Theor Appl Genet 98:1320–1328
Huelsenbeck J, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Clark L, Zhang W, Wendel J (1995) A phylogeny of the grass family (Poaceae) based on ndhF sequence data. Syst Bot 20:436–460
Crepet W, Feldman G (1991) The earliest remains of grasses in the fossil record. Am J Bot 78:1010–1014
Ehrendorfer F, Manen J, Natali A (1994) cpDNA intergene sequences corroborate restriction site data for reconstructing Rubiaceae phylogeny. Plant Syst Evol 190:245–248
Manen J, Savolainen V, Simone P (1994) The atpB and rbcL promoters in plastid DNAs of a wide dicot range. J Mol Evol 38:577–582
Golenberg EM, Clegg MT, Durbin ML (1993) Evolution of a noncoding region of the chloroplast genome. Mol Phylogenet Evol 2:52–64
Manen J, Natali A, Ehrendorfer F (1994) Phylogeny of Rubiaceae–Rubieae inferred from the sequence of a cpDNA intergene region. Plant Syst Evol 190:195–211
Savolainen V, Spichiger R, Manen J (1997) Polyphyletism of Celastrales deduced from a chloroplast noncoding DNA region. Mol Phylogenet Evol 7:145–157
Borsch T, Quandt D (2009) Mutational dynamics and phylogenetic utility of noncoding chloroplast DNA. Plant Syst Evol 282:169–199
Olmstead R, Sweere J, Wolfe K (1993) Ninety extra nucleotide in ndhF gene of tobacco chloroplast DNA: a summary of revisions to the 1986 genome sequence. Plant Mol Biol 22:1191–1193
Sugiura M (1989) The chloroplast chromosomes in land plants. Annu Rev Cell Biol 5:51–70
Wolfe K (1991) Protein-coding genes in chloroplast DNA: compilation of nucleotide sequences, data base entries, and rates of molecular evolution. Academic Press, New York
Smith J, Carroll C (1997) Phylogenetic relationships of the Episcieae (Gesneriaceae) based on ndhF sequences. Syst Bot 22:713–724
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Thompson J, Gibson T, Plewniak F et al (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882
Swofford D (2002) PAUP: phylogenetic analysis using parsimony, version 4.0 b10. Sinauer Associates, Sunderland
Kluge A, Farris J (1969) Quantitative phyletics and the evolution of anurans. Syst Zool 18:1–32
Farris J (1989) The retention index and the rescaled consistency index. Cladistics 5:417–419
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Li H, Yang J, Li D, Mller M et al (2010) A molecular phylogenetic study of Hemsleya (Cucurbitaceae) based on ITS, rpl16, trnH-psbA, and trnL DNA sequences. Plant Syst Evol 285:23–32
Ronquist F, Huelsenbeck J (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574
Posada D, Crandall K (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14(9):817–818
Tamura K, Dudley J, Nei M (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
Linder H (1987) The evolutionary history of the Poales/Restionales: a hypothesis. Kew Bull 42:297–318
Tateoka T (1965) Porteresia, a new genus of Gramineae. Bull Nat Sci Mus, Tokyo 8:405–406
Ge S, Sang T, Lu B, Hong D (1999) Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci USA 96(25):14400–14405
Ge S, Sang T, Lu B, Hong D (2001) Phylogeny of the genus oryza as revealed by molecular approaches. Rice Genet 4:89
Hillis D, Bull J (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42:182–192
Joshi S, Gupta V, Aggarwal R et al (2000) Genetic diversity and phylogenetic relationship as revealed by inter simple sequence repeat (ISSR) polymorphism in the genus Oryza. Theor Appl Genet 100:1311–1320
Zhang S, Second G (1989) Phylogenetic analysis of the tribe Oryzeae: total chloroplast DNA restriction fragment analysis (a preliminary report). Rice Genet Newsl 6:76–80
Tateoka T (1963) Relationship between oryzeae and ehrharteae, with special reference to leaf anatomy and histology. Bot Gaz 124:264–270
Ge S, Li A, Lu BR (2002) A phylogeny of the rice tribe Oryzeae (Poaceae) based on matK sequence data. Am J Bot 89(12):1967–1972
Jacobs B, Kingston J, Jacobs L (1999) The origin of grass-dominated ecosystems. Ann Mo Bot Gard 86:590–643
Kellogg E (2001) Evolutionary history of the grasses. Plant Physiol 125(3):1198–1205
Thomasson J (1978) Epidermal patterns of the lemma in some fossil and living grasses and their phylogenetic significance. Science 199(4332):975–977
Thomasson J (1980) Archaeoleersia nebraskensis (Gramineae-Oryzeae), a new fossil grass from the late tertiary of Nebraska. Am J Bot 67:876–882
Litke R (1968) Über den Nachweis tertiarer Gramineen. Monatsberichte der Deutschen Akademie der Wissenschaften zu Berlin 10:464–471
Liu ZH, Chen SL (2010) ER regulates an evolutionarily conserved apoptosis pathway. Biochem Biophys Res Commun 400(1):34–38
Liu ZH, Sun X (2008) Coronavirus phylogeny based on base-base correlation. Int J Bioinform Res Appl 4(2):211–220
Liu ZH, Meng J, Sun X (2008) A novel feature-based method for whole genome phylogenetic analysis without alignment: application to HEV genotyping and subtyping. Biochem Biophys Res Commun 368(2):223–230
Liu ZH, Jiao D, Sun X (2005) Classifying genomic sequences by sequence feature analysis. Genomics Proteomics Bioinformatics 3(4):201–205
Acknowledgments
This work was funded by National Natural Science Foundation of China (81102746), Beijing Natural Science Foundation (5113033), Scientific Research Foundation of the State Human Resource Ministry and the Education Ministry for Returned Chinese Scholars, New Star Project of Peking Union Medical College, Youth Foundation of Peking Union Medical College, the Research Fund for the Doctoral Program of Higher Education (20111106120028), “Major Drug Discovery” major science and technology research “12nd Five-Year Plan” (2012ZX09301-002-001) China Medical Board of New York (A2009001) granted to Zhihua Liu.
Author information
Authors and Affiliations
Corresponding author
Additional information
X. Zeng, Z. Yuan and X. Tong have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zeng, X., Yuan, Z., Tong, X. et al. Phylogenetic study of Oryzoideae species and related taxa of the Poaceae based on atpB-rbcL and ndhF DNA sequences. Mol Biol Rep 39, 5737–5744 (2012). https://doi.org/10.1007/s11033-011-1383-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1383-0