Abstract
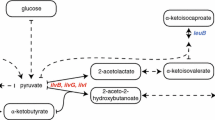
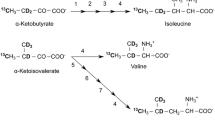
The 1H–13C HMQC signals of the 13CH3 moieties of Ile, Leu, and Val residues, in an otherwise deuterated background, exhibit narrow line-widths, and thus are useful for investigating the structures and dynamics of larger proteins. This approach, named methyl TROSY, is economical as compared to laborious methods using chemically synthesized site- and stereo-specifically isotope-labeled amino acids, such as stereo-array isotope labeling amino acids, since moderately priced, commercially available isotope-labeled α-keto acid precursors can be used to prepare the necessary protein samples. The Ile δ1-methyls can be selectively labeled, using isotope-labeled α-ketobutyrates as precursors. However, it is still difficult to prepare a residue-selectively Leu and Val labeled protein, since these residues share a common biosynthetic intermediate, α-ketoisovalerate. Another hindering drawback in using the α-ketoisovalerate precursor is the lack of stereo-selectivity for Leu and Val methyls. Here we present a differential labeling method for Leu and Val residues, using four kinds of stereo-specifically 13CH3-labeled [U–2H;15N]-leucine and -valine, which can be efficiently incorporated into a protein using Escherichia coli cellular expression. The method allows the differential labeling of Leu and Val residues with any combination of stereo-specifically isotope-labeled prochiral methyls. Since relatively small amounts of labeled leucine and valine are required to prepare the NMR samples; i.e., 2 and 10 mg/100 mL of culture for leucine and valine, respectively, with sufficient isotope incorporation efficiency, this approach will be a good alternative to the precursor methods. The feasibility of the method is demonstrated for 82 kDa malate synthase G.








Similar content being viewed by others
References
Amero C, Schanda P, Durá MA, Ayala I, Marion D, Franzetti B, Brutscher B, Boisbouvier J (2009) Fast two-dimensional NMR spectroscopy of high molecular weight protein assemblies. J Am Chem Soc 131:3448–3449
Amero C, Asunción DM, Noirclerc-Savoye M, Perollier A, Gallet B, Plevin MJ, Vernet T, Franzetti B, Boisbouvier J (2011) A systematic mutagenesis-driven strategy for site-resolved NMR studies of supramolecular assemblies. J Biomol NMR 50:229–236
Ardá A, Jiménez C, Rodríguez J (2004) A study of polychlorinated leucine derivatives: synthesis of (2S,4S)-5,5-dichloroleucine. Tetrahedron Lett 45:3241–3243
August RA, Khan JA, Moody CM, Young DW (1992) Stereospecific synthesis of (2S,4R)-[5,5,5-2H3]-leucine. Tetrahedron Lett 33:4617–4620
Ayala I, Sounier R, Usĕ N, Gans P, Boisbouvier J (2009) An efficient protocol for the complete incorporation of methyl-protonated alanine in perdeuterated protein. J Biomol NMR 43:111–119
Charrier J, Hitchcock PB, Young DW (2004) Synthesis of (2S,3R)-[3′,3′,3′-2H3]-valine and [2S,3S]-4-fluorovaline. Org Biomol Chem 2:1310–1314
Dawadi PBS, Lugtenburg J (2013) Access to any site directed stable isotope (2H, 13C, 15N, 17O and 18O) in genetically encoded amino acids. Molecules 18:482–519
Fischer M, Kloiber K, Häusler J, Ledolter K, Konrat R, Schmid W (2007) Synthesis of a 13C-methyl-group-labeled methionine precursor as a useful tool for simplifying protein structural analysis by NMR spectroscopy. ChemBioChem 8:610–612
Gans P, Hamelin O, Sounier R, Ayala I, Dura MA, Amero CD, Noirclerc-Savoye M, Franzetti B, Plevin MJ, Boisbouvier J (2010) Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew Chem 122:2002–2006
Gardner KH, Kay LE (1997) Production and incorporation of 15N, 13C, 2H (1H-δ1 methyl) isoleucine into proteins for multidimensional NMR studies. J Am Chem Soc 119:7599–7600
Gelis I, Bonvin AMJJ, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG (2007) Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell 131:756–769
Gill ML, Palmer AG (2011) Multiplet-filtered and gradient-selected zero-quantum TROSY experiments for 13C1H3 methyl groups in proteins. J Biomol NMR 51:245–251
Godoy-Ruiz R, Guo C, Tugarinov V (2010) Alanine methyl groups as NMR probes of molecular structure and dynamics in high-molecular-weight proteins. J Am Chem Soc 132:18340–18350
Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE (1999) A robust and cost-effective method for the production of Val, Leu, Ile(δ1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol NMR 13:369–374
Guo C, Tugarinov V (2010) Selective 1H-13C NMR spectroscopy of methyl groups in residually protonated samples of large proteins. J Biomol NMR 46:127–133
Howard BR, Endrizzi JA, Remington SJ (2000) Crystal Structure of Escherichia coli Malate Synthase G complexed with magnesium and glyoxylate at 2.0 Å resolution: mechanistic implications. Biochemistry 39:3156–3168
Hu H, Namanja AT, Wong S, Chen Y (2012) Selective editing of Val and Leu methyl groups in high molecular weight protein NMR. J Biomol NMR 53:113–124
Humphrey JM, Hart JA, Chamberlin AR (1995) Efficient syntheses of diastereoselectively labeled (3S)-[4-13C]-l-valine, and regioselectively labeled [3-13CH3]-l-isoleucine hydrochlorides. Bioorg Med Chem Lett 5:1315–1320
Isaacson RL, Simpson PJ, Liu M, Cota E, Zhang X, Freemont P, Matthews S (2007) A new labeling method for methyl transverse relaxation-optimized spectroscopy NMR spectra of alanine residues. J Am Chem Soc 129:15428–15429
Janin J, Miller S, Chothia C (1988) Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol 204:155–164
Kainosho M, Güntert P (2009) SAIL–stereo-array isotope labeling. Q Rev Biophys 42:247–300
Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Ono AM, Güntert P (2006) Optimal isotope labelling for NMR protein structure determinations. Nature 440:52–57
Kelly NM, Sutherland A, Willis CL (1997) Syntheses of amino acids incorporating stable isotopes. Nat Prod Rep 14:205–219
Lichtenecker R, Ludwiczek ML, Schmid W, Konrat R (2004) Simplification of protein NOESY spectra using bioorganic precursor synthesis and NMR spectral editing. J Am Chem Soc 126:5348–5349
Mas G, Crublet E, Hamelin O, Gans P, Boisbouvier J (2013) Specific labeling and assignment strategies of valine methyl groups for the NMR studies of high molecular weight proteins. J Biomol NMR. doi:10.1007/s10858-013-9785-z
Metzler WJ, Wittekind M, Goldfarb V, Mueller L, Farmer BT (1996) Incorporation of 1H/13C/15N-{Ile, Leu, Val} into a perdeuterated, 15N-labeled protein: potential in structure determination of large proteins by NMR. J Am Chem Soc 118:6800–6801
Miyanoiri Y, Takeda M, Jee JG, Ono AM, Okuma K, Terauchi T, Kainosho M (2011) Alternative SAIL-Trp for robust aromatic signal assignment and determination of the χ2 conformation by intra-residue NOEs. J Biomol NMR 51:425–435
Miyanoiri Y, Takeda M, Kainosho M (2012) Stereo-array isotope labeling method for studying protein structure and dynamics. Adv Exp Med Biol 992:83–93
Molina I, Pellicer MT, Badia J, Aguilar J, Baldoma L (1994) Molecular characterization of Escherichia coli malate synthase G: Differentiation with the malate synthase A isoenzyme. Eur J Biochem 224:541–548
Neri D, Szyperski T, Otting G, Senn H, Wüthrich K (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional carbon-13 labeling. Biochemistry 28:7510–7516
Oba M, Ueno R, Fukuoka M, Kainosho M, Nishiyama K (1995) Synthesis of L-threo- and L-erythro-[1-13C,2,3-2H2] amino-acids: Novel probes for conformational analysis of peptide side chains. J Chem Soc Perkin Trans 1:1603–1609
Oba M, Terauchi T, Miyakawa AA, Kamo H, Nishiyama K (1998) Stereoselective deuterium-labelling of diastereotopic methyl and methylene protons of l-leucine. Tetrahedron Lett 39:1595–1598
Otten R, Chu B, Krewulak KD, Vogel HJ, Mulder FAA (2010) Comprehensive and cost-effective NMR spectroscopy of methyl groups in large proteins. J Am Chem Soc 132:2952–2960
Pervushin K, Riek R, Wider G, Wüthrich K (1997) Attenuated T 2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA 94:12366–12371
Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE (1996) Selective methyl group protonation of perdeuterated proteins. J Mol Biol 263:627–636
Shekhtman A, Ghose R, Goger M, Cowburn D (2002) NMR structure determination and investigation using a reduced proton (REDPRO) labeling strategy for proteins. FEBS Lett 524:177–182
Sheppard D, Guo C, Tugarinov V (2009a) 4D, pp. 1H–13C NMR spectroscopy for assignments of alanine methyls in large and complex protein structures. J Am Chem Soc 131:1364–1365
Sheppard D, Guo C, Tugarinov V (2009b) Methyl-detected ‘out-and-back’ NMR experiments for simultaneous assignments of Alaβ and Ileγ2 methyl groups in large proteins. J Biomol NMR 43:229–238
Skrynnikov NR, Mulder FAA, Hon B, Dahlquist FW, Kay LE (2001) Probing slow time scale dynamics at methyl-containing side chains in proteins by relaxation dispersion NMR measurements: application to methionine residues in a cavity mutant of T4 lysozyme. J Am Chem Soc 123:4556–4566
Sounier R, Blanchard L, Wu Z, Boisbouvier J (2007) High-accuracy distance measurement between remote methyls in specifically protonated proteins. J Am Chem Soc 129:472–473
Takeda M, Ikeya T, Güntert P, Kainosho M (2007) Automated structure determination of proteins with the SAIL-FLYA NMR method. Nat Protoc 2:2896–2902
Takeda M, Hallenga K, Shigezane M, Waelchli M, Löhr F, Markley JL, Kainosho M (2011) Construction and performance of an NMR tube with a sample cavity formed within magnetic susceptibility matched glass. J Magn Reson 209:167–173
Takeda M, Terauchi T, Kainosho M (2012) Conformational analysis by quantitative NOE measurements of the β-proton pairs across individual disulfide bonds in proteins. J Biomol NMR 52:127–139
Torizawa T, Shimizu M, Taoka M, Miyano H, Kainosho M (2004) Efficient production of isotopically labeled proteins by cell-free synthesis: a practical protocol. J Biomol NMR 30:311–325
Tugarinov V, Kay LE (2003) Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc 125:13868–13878
Tugarinov V, Kay LE (2004a) An isotope labeling strategy for methyl TROSY spectroscopy. J Biomol NMR 28:165–172
Tugarinov V, Kay LE (2004b) Stereospecific NMR assignments of prochiral methyls, rotameric states and dynamics of valine residues in malate synthase G. J Am Chem Soc 126:9827–9836
Tugarinov V, Kay LE (2005) Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. ChemBioChem 6:1567–1577
Tugarinov V, Muhandiram R, Ayed A, Kay LE (2002) Four-dimensional NMR spectroscopy of a 723-residue protein: Chemical shift assignments and secondary structure of malate synthase G. J Am Chem Soc 124:10025–10035
Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE (2003) Cross-correlated relaxation enhanced 1H-13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc 125:10420–10428
Tugarinov V, Kanelis V, Kay LE (2006) Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc 1:749–754
Velyvis A, Kay LE (2013) Measurement of active site ionization equilibria in the 670 kDa Proteasome core particle using methyl-TROSY NMR. J Am Chem Soc 135:9259–9262
Velyvis A, Ruschak AM, Kay LE (2012) An economical method for production of 2H,13CH3-threonine for solution NMR studies of large protein complexes: Application to the 670 kDa proteasome. PLoS ONE 7:e43725
Xu Y, Matthews S (2013) TROSY NMR spectroscopy of large soluble proteins. Top Curr Chem 335:97–119
Acknowledgments
This work was supported by Platform for Drug Discovery, Informatics, and Structural Life Science (MEXT), and also in part by the Targeted Protein Research Program (MEXT). Y. M. was supported by JSPS KAKENHI Grants-in-Aid for Young Scientists (B) (23770111 and 25840021), and M. T. was supported by a JSPS KAKENHI Grant-in-Aid for Young Scientists (B) (23770109), a Grant-in-Aid for Scientific Research (C) (25440018), and a grant from the Kurata Memorial Hitachi Science and Technology Foundation. We would like to thank Dr. Tugarinov, of the University of Maryland, for providing the MSG gene and his advice regarding MSG protein production. We also thank Dr. Jérôme Boisbouvier, of Institute de Biologie Structurale Jean-Pierre Ebel, for valuable discussions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Miyanoiri, Y., Takeda, M., Okuma, K. et al. Differential isotope-labeling for Leu and Val residues in a protein by E. coli cellular expression using stereo-specifically methyl labeled amino acids. J Biomol NMR 57, 237–249 (2013). https://doi.org/10.1007/s10858-013-9784-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-013-9784-0