Abstract
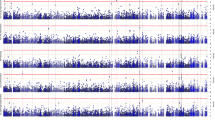
Apart from amyloid β deposition and tau neurofibrillary tangles, Alzheimer's disease (AD) is a neurodegenerative disorder characterized by neuronal loss and astrocytosis in the cerebral cortex. The goal of this study is to investigate genetic factors associated with the neuronal proportion in health and disease. To identify cell-autonomous genetic variants associated with neuronal proportion in cortical tissues, we inferred cellular population structure from bulk RNA-Seq derived from 1536 individuals. We identified the variant rs1990621 located in the TMEM106B gene region as significantly associated with neuronal proportion (p value = 6.40 × 10−07) and replicated this finding in an independent dataset (p value = 7.41 × 10−04) surpassing the genome-wide threshold in the meta-analysis (p value = 9.42 × 10−09). This variant is in high LD with the TMEM106B non-synonymous variant p.T185S (rs3173615; r2 = 0.98) which was previously identified as a protective variant for frontotemporal lobar degeneration (FTLD). We stratified the samples by disease status, and discovered that this variant modulates neuronal proportion not only in AD cases, but also several neurodegenerative diseases and in elderly cognitively healthy controls. Furthermore, we did not find a significant association in younger controls or schizophrenia patients, suggesting that this variant might increase neuronal survival or confer resilience to the neurodegenerative process. The single variant and gene-based analyses also identified an overall genetic association between neuronal proportion, AD and FTLD risk. These results suggest that common pathways are implicated in these neurodegenerative diseases, that implicate neuronal survival. In summary, we identified a protective variant in the TMEM106B gene that may have a neuronal protection effect against general aging, independent of disease status, which could help elucidate the relationship between aging and neuronal survival in the presence or absence of neurodegenerative disorders. Our findings suggest that TMEM106B could be a potential target for neuronal protection therapies to ameliorate cognitive and functional deficits.



Similar content being viewed by others
Data availability
The meta-analysis results can be accessed interactively through our Online Neurodegenerative Trait Integrative Multi-Omics Explorer (ONTIME): http://ngi.pub:5000/pheno/Neuron_QTL. Knight ADRC: https://www.synapse.org/#!Synapse:syn12181323. According to the data request terms, DIAN data are available upon request: http://dian.wustl.edu. Mayo: https://www.synapse.org/#!Synapse:syn5550404. MSSM: https://www.synapse.org/#!Synapse:syn3157743. ROSMAP: https://www.synapse.org/#!Synapse:syn3219045. CommonMind: https://www.synapse.org/#!Synapse:syn2759792. GTEx: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v7.p2.
References
Allen M, Carrasquillo MM, Funk C, Heavner BD, Zou F, Younkin CS et al (2016) Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci Data 3:160089. https://doi.org/10.1038/sdata.2016.89
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Available: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J et al (1997) Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol Histopathol 12:25–31
Askanas V, Engel WK (2001) Inclusion-body myositis: newest concepts of pathogenesis and relation to aging and Alzheimer disease. J Neuropathol Exp Neurol 60:1–14
Bateman RJ, Xiong C, Benzinger TL, Fagan AM, Goate A, Fox NC et al (2012) Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med 367:795–804
Battle A, Brown CD, Engelhardt BE, Montgomery SB (2017) Genetic effects on gene expression across human tissues. Nature 550:204–213. https://doi.org/10.1038/nature24277
Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS (2012) Overview and findings from the religious orders study. Curr Alzheimer Res 9:628–645
Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson RS (2012) Overview and findings from the rush memory and aging project. Curr Alzheimer Res 9:646–663
Brady OA, Zheng Y, Murphy K, Huang M, Hu F (2013) The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum Mol Genet 22:685–695. https://doi.org/10.1093/hmg/dds475
Broad Institute (2017) Picard: A set of command line tools for manipulating high-throughput sequencing data. Available: https://broadinstitute.github.io/picard/
Carmona-Gutierrez D, Hughes AL, Madeo F, Ruckenstuhl C (2016) The crucial impact of lysosomes in aging and longevity. Ageing Res Rev 32:2–12
Chailangkarn T, Trujillo CA, Freitas BC, Hrvoj-Mihic B, Herai RH, Yu DX et al (2016) A human neurodevelopmental model for Williams syndrome. Nature 536:338–343. https://doi.org/10.1038/nature19067
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7
Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L et al (2012) TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci 32:11213–11227. https://doi.org/10.1523/jneurosci.0521-12.2012
Consortium G (2013) The genotype-tissue expression (GTEx) project. Nat Genet 45:580–585. https://doi.org/10.1038/ng.2653
Cruchaga C, Graff C, Chiang HH, Wang J, Hinrichs AL, Spiegel N et al (2011) Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol 68:581–586. https://doi.org/10.1001/archneurol.2010.350
Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM et al (2013) GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron 78:256–268
de Leeuw CA, Mooij JM, Heskes T, Posthuma D (2015) MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11:e1004219. https://doi.org/10.1371/journal.pcbi.1004219
Delaneau O, Marchini J (2014) Integrating sequence and array data to create an improved 1000 genomes project haplotype reference panel. Nat Commun 5:3934. https://doi.org/10.1038/ncomms4934
Deming Y, Filipello F, Cignarella F, Cantoni C, Hsu S, Mikesell R et al (2018) The MS4A gene cluster is a key regulator of soluble TREM2 and Alzheimer disease risk. bioRxiv. https://doi.org/10.1101/352179
Deming Y, Li Z, Kapoor M, Harari O, Del-Aguila JL, Black K et al (2017) Genome-wide association study identifies four novel loci associated with Alzheimer’s endophenotypes and disease modifiers. Acta Neuropathol 133:839–856
Deming Y, Xia J, Cai Y, Lord J, Del-Aguila JL, Fernandez MV et al (2016) Genetic studies of plasma analytes identify novel potential biomarkers for several complex traits. Sci Rep. https://doi.org/10.1038/srep18092
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Duan J, Sanders AR, Moy W, Drigalenko EI, Brown EC, Freda J et al (2015) Transcriptome outlier analysis implicates schizophrenia susceptibility genes and enriches putatively functional rare genetic variants. Hum Mol Genet 24:4674–4685. https://doi.org/10.1093/hmg/ddv199
Espay AJ, Vizcarra JA, Marsili L, Lang AE, Simon DK, Merola A et al (2019) Revisiting protein aggregation as pathogenic in sporadic Parkinson and Alzheimer diseases. Neurology 92:329–337. https://doi.org/10.1212/wnl.0000000000006926
Fagan AM, Roe CM, Xiong C, Mintun MA, Morris JC, Holtzman DM (2007) Cerebrospinal fluid tau/beta-amyloid(42) ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol 64:343–349. https://doi.org/10.1001/archneur.64.3.noc60123
Ferrari R, Hernandez DG, Nalls MA, Rohrer JD, Ramasamy A, Kwok JB et al (2014) Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol 13:686–699. https://doi.org/10.1016/s1474-4422(14)70065-1
Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM et al (2007) Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol 179:485–500. https://doi.org/10.1083/jcb.200702115
Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M et al (2011) TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology 76:467–474. https://doi.org/10.1212/WNL.0b013e31820a0e3b
Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM et al (2016) Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 19:1442–1453. https://doi.org/10.1038/nn.4399
Gallagher MD, Posavi M, Huang P, Unger TL, Berlyand Y, Gruenewald AL et al (2017) A dementia-associated risk variant near TMEM106B alters chromatin architecture and gene expression. Am J Hum Genet 101:643–663. https://doi.org/10.1016/j.ajhg.2017.09.004
Garcia-Osta A, Cuadrado-Tejedor M, Garcia-Barroso C, Oyarzabal J, Franco R (2012) Phosphodiesterases as therapeutic targets for Alzheimer’s disease. ACS Chem Neurosci 3:832–844. https://doi.org/10.1021/cn3000907
Gaujoux R, Seoighe C (2013) Cell Mix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics 29:2211–2212. https://doi.org/10.1093/bioinformatics/btt351
Han B, Eskin E (2012) Interpreting meta-analyses of genome-wide association studies. PLoS Genet 8:e1002555. https://doi.org/10.1371/journal.pgen.1002555
Harari O, Cruchaga C, Kauwe JS, Ainscough BJ, Bales K, Pickering EH et al (2014) Phosphorylated tau-Aβ42 ratio as a continuous trait for biomarker discovery for early-stage Alzheimer’s disease in multiplex immunoassay panels of cerebrospinal fluid. Biol Psychiatry 75:723–731. https://doi.org/10.1016/j.biopsych.2013.11.032
Hardy J, Selkoe DJ (2002) The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297:353–356
Holtzman DM, Morris JC, Goate AM (2011) Alzheimer’s disease: the challenge of the second century. Sci Transl Med 3:77sr71
Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR (2012) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44:955–959. https://doi.org/10.1038/ng.2354
Infante J, Llorca J, Rodero L, Palacio E, Berciano J, Combarros O (2002) Polymorphism at codon 174 of the prion-like protein gene is not associated with sporadic Alzheimer’s disease. Neurosci Lett 332:213–215
Jentsch TJ (2007) Chloride and the endosomal-lysosomal pathway: emerging roles of CLC chloride transporters. J Physiol 578:633–640. https://doi.org/10.1113/jphysiol.2006.124719
Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC et al (2016) A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry 21:108–117. https://doi.org/10.1038/mp.2015.23
Kauwe JS, Bailey MH, Ridge PG, Perry R, Wadsworth ME, Hoyt KL et al (2014) Genome-wide association study of CSF levels of 59 Alzheimer’s disease candidate proteins: significant associations with proteins involved in amyloid processing and inflammation. PLoS Genet 10:e1004758
Keage HA, Hunter S, Matthews FE, Ince PG, Hodges J, Hokkanen SR et al (2014) TDP-43 pathology in the population: prevalence and associations with dementia and age. J Alzheimers Dis 42:641–650. https://doi.org/10.3233/jad-132351
Kelly MP (2017) A role for phosphodiesterase 11A (PDE11A) in the formation of social memories and the stabilization of mood. Adv Neurobiol 17:201–230. https://doi.org/10.1007/978-3-319-58811-7_8
Kelly MP, Adamowicz W, Bove S, Hartman AJ, Mariga A, Pathak G et al (2014) Select 3′,5′-cyclic nucleotide phosphodiesterases exhibit altered expression in the aged rodent brain. Cell Signal 26:383–397. https://doi.org/10.1016/j.cellsig.2013.10.007
Ko DC, Milenkovic L, Beier SM, Manuel H, Buchanan J, Scott MP (2005) Cell-autonomous death of cerebellar purkinje neurons with autophagy in Niemann-Pick type C disease. PLoS Genet 1:81–95. https://doi.org/10.1371/journal.pgen.0010007
Koike M, Shibata M, Ohsawa Y, Nakanishi H, Koga T, Kametaka S et al (2003) Involvement of two different cell death pathways in retinal atrophy of cathepsin D-deficient mice. Mol Cell Neurosci 22:146–161
Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E et al (2005) Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease). Am J Pathol 167:1713–1728. https://doi.org/10.1016/s0002-9440(10)61253-9
Kushima I, Aleksic B, Nakatochi M, Shimamura T, Okada T, Uno Y et al (2018) Comparative analyses of copy-number variation in autism spectrum disorder and schizophrenia reveal etiological overlap and biological insights. Cell Rep 24:2838–2856. https://doi.org/10.1016/j.celrep.2018.08.022
Kypri E, Schmauch C, Maniak M, De Lozanne A (2007) The BEACH protein LvsB is localized on lysosomes and postlysosomes and limits their fusion with early endosomes. Traffic (Copenhagen, Denmark) 8:774–783. https://doi.org/10.1111/j.1600-0854.2007.00567.x
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45:1452–1458. https://doi.org/10.1038/ng.2802
Lang CM, Fellerer K, Schwenk BM, Kuhn PH, Kremmer E, Edbauer D et al (2012) Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J Biol Chem 287:19355–19365. https://doi.org/10.1074/jbc.M112.365098
Laszlo L, Lowe J, Self T, Kenward N, Landon M, McBride T et al (1992) Lysosomes as key organelles in the pathogenesis of prion encephalopathies. J Pathol 166:333–341. https://doi.org/10.1002/path.1711660404
Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB (2007) ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol 17:1561–1567. https://doi.org/10.1016/j.cub.2007.07.029
Leidal AM, Levine B, Debnath J (2018) Autophagy and the cell biology of age-related disease. Nat Cell Biol 20:1338–1348. https://doi.org/10.1038/s41556-018-0235-8
Li Z, Del-Aguila JL, Dube U, Budde J, Martinez R, Black K et al (2018) Genetic variants associated with Alzheimer’s disease confer different cerebral cortex cell-type population structure. Genome Med 10:43. https://doi.org/10.1186/s13073-018-0551-4
Murray ME, Cannon A, Graff-Radford NR, Liesinger AM, Rutherford NJ, Ross OA et al (2014) Differential clinicopathologic and genetic features of late-onset amnestic dementias. Acta Neuropathol 128:411–421. https://doi.org/10.1007/s00401-014-1302-2
Murray ME, Dickson DW (2014) Is pathological aging a successful resistance against amyloid-beta or preclinical Alzheimer’s disease? Alzheimers Res Ther 6:24. https://doi.org/10.1186/alzrt254
Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F et al (2019) Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 37:773–782. https://doi.org/10.1038/s41587-019-0114-2
Nicholson AM, Finch NA, Wojtas A, Baker MC, Perkerson RB 3rd, Castanedes-Casey M et al (2013) TMEM106B p. T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J Neurochem 126:781–791. https://doi.org/10.1111/jnc.12329
Nixon RA (2005) Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol Aging 26:373–382. https://doi.org/10.1016/j.neurobiolaging.2004.09.018
Nixon RA, Yang DS, Lee JH (2008) Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy 4:590–599
Pacheco CD, Kunkel R, Lieberman AP (2007) Autophagy in Niemann-Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum Mol Genet 16:1495–1503. https://doi.org/10.1093/hmg/ddm100
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. https://doi.org/10.1038/nmeth.4197
Piccio L, Deming Y, Del-Aguila JL, Ghezzi L, Holtzman DM, Fagan AM et al (2016) Cerebrospinal fluid soluble TREM2 is higher in Alzheimer disease and associated with mutation status. Acta Neuropathol 131:925–933. https://doi.org/10.1007/s00401-016-1533-5
Premi E, Grassi M, van Swieten J, Galimberti D, Graff C, Masellis M et al (2017) Cognitive reserve and TMEM106B genotype modulate brain damage in presymptomatic frontotemporal dementia: a GENFI study. Brain 140:1784–1791. https://doi.org/10.1093/brain/awx103
Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R et al (2014) Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 17:1418–1428. https://doi.org/10.1038/nn.3801
Reid E, Connell J, Edwards TL, Duley S, Brown SE, Sanderson CM (2005) The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum Mol Genet 14:19–38. https://doi.org/10.1093/hmg/ddi003
Rhinn H, Abeliovich A (2017) Differential aging analysis in human cerebral cortex identifies variants in TMEM106B and GRN that regulate aging phenotypes. Cell Syst 4:404–415.e5. https://doi.org/10.1016/j.cels.2017.02.009
Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC (2007) Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem 282:5641–5652. https://doi.org/10.1074/jbc.M609532200
Schroder B, Franz B, Hempfling P, Selbert M, Jurgens T, Kretzschmar HA et al (2001) Polymorphisms within the prion-like protein gene (Prnd) and their implications in human prion diseases, Alzheimer’s disease and other neurological disorders. Hum Genet 109:319–325. https://doi.org/10.1007/s004390100591
Schwenk BM, Lang CM, Hogl S, Tahirovic S, Orozco D, Rentzsch K et al (2014) The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. EMBO J 33:450–467. https://doi.org/10.1002/embj.201385857
Shirk AJ, Anderson SK, Hashemi SH, Chance PF, Bennett CL (2005) SIMPLE interacts with NEDD4 and TSG101: evidence for a role in lysosomal sorting and implications for Charcot–Marie–Tooth disease. J Neurosci Res 82:43–50. https://doi.org/10.1002/jnr.20628
Stagi M, Klein ZA, Gould TJ, Bewersdorf J, Strittmatter SM (2014) Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol Cell Neurosci 61:226–240. https://doi.org/10.1016/j.mcn.2014.07.006
Suárez-Calvet M, Capell A, Caballero MÁA, Morenas-Rodríguez E, Fellerer K, Franzmeier N et al (2018) CSF progranulin increases in the course of Alzheimer’s disease and is associated with sTREM2, neurodegeneration and cognitive decline. EMBO Mol Med 10:e9712
Sul JH, Han B, Ye C, Choi T, Eskin E (2013) Effectively identifying eQTLs from multiple tissues by combining mixed model and meta-analytic approaches. PLoS Genet 9:e1003491. https://doi.org/10.1371/journal.pgen.1003491
Toledo JB, Cairns NJ, Da X, Chen K, Carter D, Fleisher A et al (2013) Clinical and multimodal biomarker correlates of ADNI neuropathological findings. Acta Neuropathol Commun 1:65. https://doi.org/10.1186/2051-5960-1-65
Turner SD (2014) qqman: an R package for visualizing GWAS results using Q–Q and manhattan plots. bioRxiv. https://doi.org/10.1101/005165
Uchino A, Takao M, Hatsuta H, Sumikura H, Nakano Y, Nogami A et al (2015) Incidence and extent of TDP-43 accumulation in aging human brain. Acta Neuropathol Commun 3:35. https://doi.org/10.1186/s40478-015-0215-1
Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR et al (2010) Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet 42:234–239. https://doi.org/10.1038/ng.536
Vass R, Ashbridge E, Geser F, Hu WT, Grossman M, Clay-Falcone D et al (2011) Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol 121:373–380. https://doi.org/10.1007/s00401-010-0782-y
Vonsattel JP, DiFiglia M (1998) Huntington disease. J Neuropathol Exp Neurol 57:369–384
Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP et al (2018) Comprehensive functional genomic resource and integrative model for the human brain. Science. https://doi.org/10.1126/science.aat8464
Wang L, Nie J, Sicotte H, Li Y, Eckel-Passow JE, Dasari S et al (2016) Measure transcript integrity using RNA-seq data. BMC Bioinform 17:58. https://doi.org/10.1186/s12859-016-0922-z
Wang M, Beckmann ND, Roussos P, Wang E, Zhou X, Wang Q et al (2018) The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci Data 5:180185. https://doi.org/10.1038/sdata.2018.185
Watanabe K, Taskesen E, van Bochoven A, Posthuma D (2017) Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8:1826. https://doi.org/10.1038/s41467-017-01261-5
Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC (2003) Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem 278:25009–25013. https://doi.org/10.1074/jbc.M300227200
Wennberg AM, Whitwell JL, Tosakulwong N, Weigand SD, Murray ME, Machulda MM et al (2019) The influence of tau, amyloid, alpha-synuclein, TDP-43, and vascular pathology in clinically normal elderly individuals. Neurobiol Aging 77:26–36. https://doi.org/10.1016/j.neurobiolaging.2019.01.008
White CC, Yang HS, Yu L, Chibnik LB, Dawe RJ, Yang J et al (2017) Identification of genes associated with dissociation of cognitive performance and neuropathological burden: multistep analysis of genetic, epigenetic, and transcriptional data. PLoS Med 14:e1002287. https://doi.org/10.1371/journal.pmed.1002287
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82. https://doi.org/10.1016/j.ajhg.2010.11.011
Zhou X, Sun L, Brady OA, Murphy KA, Hu F (2017) Elevated TMEM106B levels exaggerate lipofuscin accumulation and lysosomal dysfunction in aged mice with progranulin deficiency. Acta Neuropathol Commun 5:9. https://doi.org/10.1186/s40478-017-0412-1
Acknowledgements
We thank all the participants and their families, as well as the many institutions and their staff that provided support for the studies involved in this collaboration. We also thank Dr. Jae Hoon Sul for his help with the Meta-Tissue analysis applied in this study.
Funding
This work was supported by Grants from the National Institutes of Health (R01AG044546, P01AG003991, RF1AG053303, R01AG058501, U01AG058922, R01AG057777, and U01AG058922), the Alzheimer’s Association (NIRG-11-200110, BAND-14-338165, AARG-16-441560 and BFG-15-362540). BAB is supported by 2018 pilot funding from the Hope Center for Neurological Disorders and the Danforth Foundation Challenge at Washington University. The recruitment and clinical characterization of research participants at Washington University were supported by NIH P50 AG05681, P01 AG03991, and P01 AG026276. This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders and the Departments of Neurology and Psychiatry at Washington University School of Medicine. DIAN: Data collection and sharing for this project was supported by The Dominantly Inherited Alzheimer’s Network (DIAN, UF1AG032438) funded by the National Institute on Aging (NIA), the German Center for Neurodegenerative Diseases (DZNE), and the Raul Carrea Institute for Neurological Research (FLENI). Partial support was provided by the Research and Development Grants for Dementia from the Japan Agency for Medical Research and Development, AMED, and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI). This manuscript has been reviewed by DIAN Study investigators for scientific content and consistency of data interpretation with previous DIAN Study publications. We acknowledge the altruism of the participants and their families and contributions of the DIAN research and support staff at each of the participating sites for their contributions to this study. Mayo RNAseq: Study data were provided by the following sources: The Mayo Clinic Alzheimer’s Disease Genetic Studies, led by Dr. Nilufer Taner and Dr. Steven G. Younkin, Mayo Clinic, Jacksonville, FL using samples from the Mayo Clinic Study of Aging, the Mayo Clinic Alzheimer’s Disease Research Center, and the Mayo Clinic Brain Bank. Data collection was supported through funding by NIA Grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216, R01 AG003949, NINDS grant R01 NS080820, CurePSP Foundation, and support from Mayo Foundation. Study data includes samples collected through the Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona. The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research. MSBB: These data were generated from postmortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank and were provided by Dr. Eric Schadt from Mount Sinai School of Medicine.
Author information
Authors and Affiliations
Contributions
ZL analyzed the data and wrote the manuscript. FGF performed the genotype QC, imputation, and merging. ZL, UB, JLD, KAM, JPB, OH, CC performed the RNA-Seq data preprocessing and QC. ZL, UB, JLD, KAM, JPB, FW, OH, CC performed the public data assembly and phenotype QC. ZL, FGF, UD, JLD, KAM, MVF, LI, JPB, AML, YD, JP, CY, JAB contributed to the analysis pipeline applied in the study. ZL, FGF, UD, JLD, KAM, MVF, LI, JPB, YD, JP, CY, JAB, QW, BAB, JDD, OH, CC contributed to the result interpretation. JLB, RD, KB, JPB, BAB contributed to the genotype and RNA-Seq data generation. JCM and PJP provided the Knight ADRC data. CC, OH, JDD, ZL designed the study. CC supervised the project. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
CC receives research support from: Biogen, EISAI, Alector and Parabon. The funders of the study had no role in the collection, analysis, interpretation of data; or in the writing of the report; or in the decision to submit the paper for publication. CC is a member of the advisory board of ADx Healthcare and Vivid Genomics.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Z., Farias, F.H.G., Dube, U. et al. The TMEM106B FTLD-protective variant, rs1990621, is also associated with increased neuronal proportion. Acta Neuropathol 139, 45–61 (2020). https://doi.org/10.1007/s00401-019-02066-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-019-02066-0