Abstract
A ranked tree topology is a tree topology with a temporal ordering of its coalescence events. Under the multispecies coalescent model, we consider ranked gene tree topologies realized along the branches of ranked species trees, where one gene copy is sampled for each species. Previous results have demonstrated that for almost all ranked species tree topologies with at least five species, there exists a set of branch lengths such that the maximally probable ranked gene tree topologies—those generated with the highest probability under the model—do not match the species tree ranked topology. Here, we focus on the agreement of a ranked species tree with its maximally probable ranked gene tree topologies in terms of their unranked topology, that is, disregarding the ordering of the coalescence events. We show that although the set of maximally probable ranked gene tree topologies for a ranked species tree can contain ranked trees with different unranked topologies, at least one of these maximal ranked gene tree topologies must have the same unranked topology as the species tree. Our results contribute to the study of the relationships between gene trees and species trees.





Similar content being viewed by others
References
Degnan JH, Rosenberg NA (2006) Discordance of species trees with their most likely gene trees. PLoS Genet. 2:762–768
Degnan JH, Rosenberg NA (2009) Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24:332–340
Degnan JH, Salter LA (2005) Gene tree distributions under the coalescent process. Evolution 59:24–37
Degnan JH, DeGiorgio M, Bryant D, Rosenberg NA (2009) Properties of consensus methods for inferring species trees from gene trees. Syst. Biol. 58:35–54
Degnan JH, Rosenberg NA, Stadler T (2012a) A characterization of the set of species trees that produce anomalous ranked gene trees. IEEE/ACM Trans. Comput. Biol. Bioinform. 9:1558–1568
Degnan JH, Rosenberg NA, Stadler T (2012b) The probability distribution of ranked gene trees on a species tree. Math. Biosci. 235:45–55
Disanto F, Rosenberg NA (2014) On the number of ranked species trees producing anomalous ranked gene trees. IEEE/ACM Trans. Comput. Biol. Bioinform. 11:1229–1238
Ewing GB, Ebersberger I, Schmidt HA, von Haeseler A (2008) Rooted triple consensus and anomalous gene trees. BMC Evol. Biol. 8:118
Felsenstein J (2004) Inferring phylogenies. Sinauer, Sunderland
Heled J, Drummond AJ (2010) Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27:570–580
Hudson RR (1983) Testing the constant-rate neutral allele model with protein sequence data. Evolution 37:203–217
Kubatko LS, Carstens BC, Knowles LL (2009) Stem: species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics 25:971–973
Liu L, Pearl DK (2007) Species trees from gene trees: reconstructing Bayesian posterior distributions of a species phylogeny using estimated gene tree distributions. Syst. Biol. 56:504–514
Liu L, Yu L (2011) Estimating species trees from unrooted gene trees. Syst. Biol. 60:661–667
Liu L, Yu L, Pearl DK, Edwards SV (2009) Estimating species phylogenies using coalescence times among sequences. Syst. Biol. 58:468–477
Liu L, Yu L, Pearl DK (2010) Maximum tree: a consistent estimator of the species tree. J. Math. Biol. 60:95–106
Maddison WP (1997) Gene trees in species trees. Syst. Biol. 46:523–536
Maddison WP, Knowles LL (2006) Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 55:21–30
Mossel E, Roch S (2010) Incomplete lineage sorting: consistent phylogeny estimation from multiple loci. IEEE/ACM Trans. Comput. Biol. Bioinform. 7:166–171
Pamilo P, Nei M (1988) Relationships between gene trees and species trees. Mol. Biol. Evol. 5:568–583
Rosenberg NA (2002) The probability of topological concordance of gene trees and species trees. Theor. Popul. Biol. 61:225–247
Rosenberg NA (2006) The mean and variance of the numbers of \(r\)-pronged nodes and \(r\)-caterpillars in Yule-generated genealogical trees. Ann. Comb. 10:129–146
Stadler T, Degnan JH (2012) A polynomial time algorithm for calculating the probability of a ranked gene tree given a species tree. Algorithms Mol. Biol. 7:7
Than C, Nakhleh L (2009) Species tree inference by minimizing deep coalescences. PLoS Comput. Biol. 5:e1000501
Wu Y (2012) Coalescent-based species tree inference from gene tree topologies under incomplete lineage sorting by maximum likelihood. Evolution 66:763–775
Acknowledgements
We thank Noah A. Rosenberg for discussions. Support was provided by a Rita Levi-Montalcini Grant to FD from the Ministero dell’Istruzione, dell’Università e della Ricerca.
Author information
Authors and Affiliations
Contributions
FD: wrote the paper. FD, PM, and GN: designed the study. PM and GN: derived theorems. PM and GN: implemented software.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix 1. Proof of the existence of \(\beta \)
Appendix 1. Proof of the existence of \(\beta \)
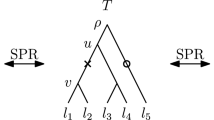
We demonstrate the existence of a point \(\beta \) of G as defined in step (ii) of the construction described in Sect. 3.1.1. In particular, for a fixed realization of G in S, we seek a point \(\beta \ne \gamma _{i^*}\) contemporary with \(\gamma _{i^*}\), i.e. at the same height of \(\gamma _{i^*}\) in S, lying on a lineage of G that has all its descending taxa below the branch \(\alpha \) of S determined in step (i) of the same construction.
First, observe that the node \(\gamma _{i^*}\) of G cannot have any of its descending taxa placed below \(\alpha \). Indeed, from step (i), \(\alpha \) is a branch of \((S_{|G_{i^*}})_{\ell }\) external to \(S_{|(G_{i^*})_{\ell }}\)—in fact, \(\alpha \) coalesces with the root branch of \(S_{|(G_{i^*})_{\ell }}\) (Fig. 4). Hence, none of the lineages of \(G_{i^*}\) passes through \(\alpha \).
Second, with respect to the considered realization of G in S, take any lineage (branch) \(\rho \) of G containing a point p contemporary with \(\gamma _{i^*}\) and such that at least one of the taxa descending from \(\rho \) is placed below the branch \(\alpha \). Such a lineage must clearly exists and, as shown above, it cannot contain the node \(\gamma _{i^*}\) of G, i.e. \(p \ne \gamma _{i^*}\). We claim that p has all its descending taxa below \(\alpha \), and thus it satisfies the definition of \(\beta \). Suppose a contrario that from p descends a taxon of G that is not placed below \(\alpha \). Since p has also a descending taxon below \(\alpha \), there must be at least one internal node of G descending from p. Let \(\gamma _v\) be the most ancient internal node of G descending from p, where \(v > i^*\). As for the point p, the node \(\gamma _v\) has some descending taxa below \(\alpha \) and some others that are not placed below \(\alpha \). Hence, since \(\alpha \) is the branch of S that coalesces with the root branch of \(S_{| ( G_{i^*} )_{\ell } }\) (Fig. 4), \([S_{|G_v}]\) strictly contains \([S_{| ( G_{i^*} )_{\ell } }]\). Note that \([S_{|G_v}] = [G_v]\), as \(v > i^*\), and \([S_{| ( G_{i^*} )_{\ell } }] = [(G_{i^*} )_{\ell }]\). Therefore, the set of taxa of \(G_v\) strictly contains the set of taxa of \((G_{i^*} )_{\ell }\), which implies that \(\gamma _v\) must be equal or ancestral to \(\gamma _{i^*}\) in G. This is in contrast with \(v > i^*\). \(\Box \)
Rights and permissions
About this article
Cite this article
Disanto, F., Miglionico, P. & Narduzzi, G. On the unranked topology of maximally probable ranked gene tree topologies. J. Math. Biol. 79, 1205–1225 (2019). https://doi.org/10.1007/s00285-019-01392-x
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-019-01392-x