Abstract
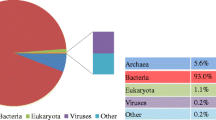
This study applied Illumina high-throughput sequencing to explore the microbial communities and functions in anaerobic digestion sludge (ADS) from two wastewater treatment plants based on a metagenomic view. Taxonomic analysis using SILVA SSU database indicated that Proteobacteria (9.52–13.50 %), Bacteroidetes (7.18 %–10.65 %) and Firmicutes (7.53 %–9.46 %) were the most abundant phyla in the ADS. Differences of microbial communities between the two types of ADS were identified. Genera of Methanosaeta and Methanosarcina were the major methanogens. Functional analysis by SEED subsystems showed that the basic metabolic functions of metagenomes in the four ADS samples had no significant difference among them, but they were different from other microbial communities from activated sludge, human faeces, ocean and soil. Abundances of genes in methanogenesis pathway were also quantified using a methanogenesis genes database extracted from KEGG. Results showed that acetotrophic was the major methanogenic pathway in the anaerobic sludge digestion.




Similar content being viewed by others
References
Albertsen M, Hansen LBS, Saunders AM, Nielsen PH, Nielsen KL (2011) A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J 6(6):1094–1106. doi:10.1038/ismej.2011.176
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Ariesyady H, Ito T, Okabe S (2007) Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res 41(7):1554–1568. doi:10.1016/j.watres.2006.12.036
Braguglia CM, Gagliano MC, Rossetti S (2012) High frequency ultrasound pretreatment for sludge anaerobic digestion: effect on floc structure and microbial population. Bioresour Technol 110:43–49. doi:10.1016/j.biortech.2012.01.074
Breitbart M, Hoare A, Nitti A, Siefert J, Haynes M, Dinsdale E, Edwards R, Souza V, Rohwer F, Hollander D (2009) Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro Ciénegas, Mexico. Environ Microbiol 11(1):16–34. doi:10.1111/j.1462-2920.2008.01725.x
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37(Database issue):D141–D145. doi:10.1093/nar/gkn879
Delmont TO, Prestat E, Keegan KP, Faubladier M, Robe P, Clark IM, Pelletier E, Hirsch PR, Meyer F, Gilbert JA, Le Paslier D, Simonet P, Vogel TM (2012) Structure, fluctuation and magnitude of a natural grassland soil metagenome. ISME J 6:1677–1687. doi:10.1038/ismej.2011.197
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72(7):5069–5072. doi:10.1128/AEM.03006-05
DiPippo JL, Nesbo CL, Dahle H, Doolittle WF, Birkland NK, Noll KM (2009) Kosmotoga olearia gen. nov., sp. nov., a thermophilic, anaerobic heterotroph isolated from an oil production fluid. Int J Syst Evol Micr 59(12):2991–3000. doi:10.1099/ijs.0.008045-0
Fang H, Cai L, Yu Y, Zhang T (2013) Metagenomic analysis reveals the prevalence of biodegradation genes for organic pollutants in activated sludge. Bioresour Technol 129:209–218. doi:10.1016/j.biortech.2012.11.054
Garcia SL, Jangid K, Whitman WB, Das KC (2011) Transition of microbial communities during the adaption to anaerobic digestion of carrot waste. Bioresour Technol 102(15):7249–7256. doi:10.1016/j.biortech.2011.04.098
Garcia-Peña EI, Parameswaran P, Kang DW, Canul-Chan M, Krajmalnik-Brown R (2011) Anaerobic digestion and co-digestion processes of vegetable and fruit residues: process and microbial ecology. Bioresour Technol 102(20):9447–9455. doi:10.1016/j.biortech.2011.07.068
Gomez-Alvarez V, Teal TK, Schmidt TM (2009) Systematic artifacts in metagenomes from complex microbial communities. ISME J 3(11):1314–1317. doi:10.1038/ismej.2009.72
Han Y, Liu J, Guo X, Li L (2012) Micro-environment characteristics and microbial communities in activated sludge flocs of different particle size. Bioresour Technol 124:252–258. doi:10.1016/j.biortech.2012.08.008
Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331(6016):463–467. doi:10.1126/science.1200387
Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17(3):377–386. doi:10.1101/gr.5969107
Jaenicke S, Ander C, Bekel T, Bisdorf R, Dröge M, Gartemann K-H, Jünemann S, Kaiser O, Krause L, Tille F, Zakrzewski M, Pühler A, Schlüter A, Goesmann A (2011) Comparative and joint analysis of two metagenomic datasets from a biogas fermenter obtained by 454-pyrosequencing. PLoS ONE 6(1):e14519. doi:10.1371/journal.pone.0014519
Jung JY, Lee SH, Kim JM, Park MS, Bae JW, Hahn Y, Madsen EL, Jeon CO (2011) Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl Environ Microb 77(7):2264–2274. doi:10.1128/aem.02157-10
Kim J, Park C, Kim T-H, Lee M, Kim S, Kim E-W, Lee J (2003) Effects of various pretreatments for enhanced anaerobic digestion with waste activated sludge. J Biosci Bioeng 95(3):271–275
Knight R, Jansson J, Field D, Fierer N, Desai N, Fuhrman JA, Hugenholtz P, van der Lelie D, Meyer F, Stevens R, Bailey MJ, Gordon JI, Kowalchuk GA, Gilbert JA (2012) Unlocking the potential of metagenomics through replicated experimental design. Nat Biotechnol 30(6):513–520. doi:10.1038/nbt.2235
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19(9):1639–1645. doi:10.1101/gr.092759.109
Li A, Yn C, Wang X, Ren L, Yu J, Liu X, Yan J, Zhang L, Wu S, Li S (2013) A pyrosequencing-based metagenomic study of methane-producing microbial community in solid-state biogas reactor. Biotechnol Biofuels. doi:10.1186/1754-6834-6-3
Ma LP, Li B, Zhang T (2014) Abundant rifampin resistance genes and significant correlations of antibiotic resistance genes and plasmids in various environments revealed by metagenomic analysis. Appl Microbiol Biot. doi:10.1007/s00253-014-5511-3
Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, Rubin EM, Jansson JK (2011) Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480(7377):368–371. doi:10.1038/nature10576
Mardis ER (2008) The impact of next-generation sequencing technology on genetics. Trends Genet 24(3):133–141. doi:10.1016/j.tig.2007.12.007
Metzker ML (2009) Sequencing technologies — the next generation. Nat Rev Genet 11(1):31–46. doi:10.1038/nrg2626
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA (2008) The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma 9(1):386. doi:10.1186/1471-2105-9-386
Nguyen HD, Cao B, Mishra B, Boyanov MI, Kemner KM, Fredrickson JK, Beyenal H (2012) Microscale geochemical gradients in Hanford 300 Area sediment biofilms and influence of uranium. Water Res 46(1):227–234. doi:10.1016/j.watres.2011.10.054
Noor E, Eden E, Milo R, Alon U (2010) Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol Cell 39(5):809–820. doi:10.1016/j.molcel.2010.08.031
Novak JT, Chon DH, Curtis B-A, Doyle M (2007) Biological solids reduction using the cannibal process. Water Environ Res 79(12):2380–2386. doi:10.2175/106143007x183862
Nunoura T, Hirai M, Imachi H, Miyazaki M, Makita H, Hirayama H, Furushima Y, Yamamoto H, Takai K (2010) Kosmotoga arenicorallina sp. nov. a thermophilic and obligately anaerobic heterotroph isolated from a shallow hydrothermal system occurring within a coral reef, southern part of the Yaeyama Archipelago, Japan, reclassification of Thermococcoides shengliensis as Kosmotoga shengliensis comb. nov., and emended description of the genus Kosmotoga. Arch Microbiol 192(10):811–819. doi:10.1007/s00203-010-0611-7
Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33(17):5691–5702. doi:10.1093/nar/gki866
Palatsi J, Illa J, Prenafeta-Boldú FX, Laureni M, Fernandez B, Angelidaki I, Flotats X (2010) Long-chain fatty acids inhibition and adaptation process in anaerobic thermophilic digestion: batch tests, microbial community structure and mathematical modelling. Bioresour Technol 101(7):2243–2251. doi:10.1016/j.biortech.2009.11.069
Palatsi J, Viñas M, Guivernau M, Fernandez B, Flotats X (2011) Anaerobic digestion of slaughterhouse waste: main process limitations and microbial community interactions. Bioresour Technol 102(3):2219–2227. doi:10.1016/j.biortech.2010.09.121
Papagianni M (2012) Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb Cell Fact 11(50)
Pérez-Elvira SI, Nieto Diez P, Fdz-Polanco F (2006) Sludge minimisation technologies. Rev Environ Sci Bio Technol 5(4):375–398. doi:10.1007/s11157-005-5728-9
Pfister CA, Meyer F, Antonopoulos DA (2010) Metagenomic profiling of a microbial assemblage associated with the California Mussel: a node in networks of carbon and nitrogen cycling. PLoS ONE 5(5):e10518. doi:10.1371/journal.pone.0010518
Prosser JI (2010) Replicate or lie. Environ Microbiol 12(7):1806–1810. doi:10.1111/j.1462-2920.2010.02201.x
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35(21):7188–7196. doi:10.1093/nar/gkm864
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J-M, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Antolin M, Artiguenave F, Blottiere H, Borruel N, Bruls T, Casellas F, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Forte M, Friss C, van de Guchte M, Guedon E, Haimet F, Jamet A, Juste C, Kaci G, Kleerebezem M, Knol J, Kristensen M, Layec S, Le Roux K, Leclerc M, Maguin E, Melo Minardi R, Oozeer R, Rescigno M, Sanchez N, Tims S, Torrejon T, Varela E, de Vos W, Winogradsky Y, Zoetendal E, Bork P, Ehrlich SD, Wang J (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464(7285):59–65. doi:10.1038/nature08821
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(D1):D590–D596. doi:10.1093/nar/gks1219
Rincón B, Borja R, González JM, Portillo MC, Sáiz-Jiménez C (2008) Influence of organic loading rate and hydraulic retention time on the performance, stability and microbial communities of one-stage anaerobic digestion of two-phase olive mill solid residue. Biochem Eng J 40(2):253–261. doi:10.1016/j.bej.2007.12.019
Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J, Li T, Camacho P, Sghir A (2009) Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J 3(6):700–714. doi:10.1038/ismej.2009.2
Sahlstrom L (2003) A review of survival of pathogenic bacteria in organic waste used in biogas plants. Bioresour Technol 87:161–166
Shi P, Jia S, Zhang X-X, Zhang T, Cheng S, Li A (2013) Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water. Water Res 47(1):111–120. doi:10.1016/j.watres.2012.09.046
Shin SG, Lee S, Lee C, Hwang K, Hwang S (2010) Qualitative and quantitative assessment of microbial community in batch anaerobic digestion of secondary sludge. Bioresour Technol 101(24):9461–9470. doi:10.1016/j.biortech.2010.07.081
Sun L, Randall CW, Novak JT (2010) The influence of sludge interchange times on the oxic–settling–anoxic process. Water Environ Res 82(6):519–523. doi:10.2175/106143009x12487095236711
Supaphol S, Jenkins SN, Intomo P, Waite IS, O’Donnell AG (2011) Microbial community dynamics in mesophilic anaerobic co-digestion of mixed waste. Bioresour Technol 102(5):4021–4027. doi:10.1016/j.biortech.2010.11.124
Traversi D, Villa S, Lorenzi E, Degan R, Gilli G (2012) Application of a real-time qPCR method to measure the methanogen concentration during anaerobic digestion as an indicator of biogas production capacity. J Environ Manag 111:173–177. doi:10.1016/j.jenvman.2012.07.021
Tringe SG (2005) Comparative metagenomics of microbial communities. Science 308(5721):554–557. doi:10.1126/science.1107851
Whiticar MJ (1999) Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem Geol 161(1–3):291–314
Ye L, Zhang T (2012) Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl Microbiol Biot 97(6):2381–2690. doi:10.1007/s00253-012-4082-4
Yu K, Zhang T (2013) Construction of customized sub-databases from NCBI-nr database for rapid annotation of huge metagenomic datasets using a combined BLAST and MEGAN approach. PLoS ONE 8(4):e59831. doi:10.1371/journal.pone.0059831
Yu Y, Lee C, Hwang S (2005) Analysis of community structures in anaerobic processes using a quantitative real-time PCR method. Water Sci Technol 52(1–2):85–91
Zhang T, Shao M-F, Ye L (2011) 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J 6(6):1137–1147. doi:10.1038/ismej.2011.188
Zhou H-W, Li D-F, Tam NF-Y, Jiang X-T, Zhang H, Sheng H-F, Qin J, Liu X, Zou F (2010) BIPES, a cost-effective high-throughput method for assessing microbial diversity. ISME J 5(4):741–749. doi:10.1038/ismej.2010.160
Acknowledgments
This work is supported by Hong Kong General Research Fund (7198/10E). Ying Yang, Ke Yu, and Yu Xia thank The University of Hong Kong for the postgraduate studentship. We thank the Drainage Services Department of the Government of the Hong Kong Special Administrative Region for their support for this study.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 660 kb)
Rights and permissions
About this article
Cite this article
Yang, Y., Yu, K., Xia, Y. et al. Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl Microbiol Biotechnol 98, 5709–5718 (2014). https://doi.org/10.1007/s00253-014-5648-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5648-0