Abstract
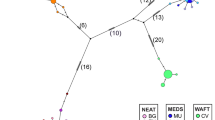
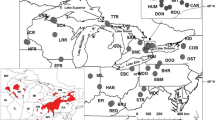
The Atlantic surfclam, Spisula solidissima (Dillwyn), is broadly distributed in sandy sediments of the western North Atlantic between the Gulf of St. Lawrence and the Gulf of Mexico. In the United States, a substantial commercial fishery between Long Island and Cape Hatteras harvests offshore populations of one subspecies, S. s. solidissima. A smaller coastal form, S. s. similis Say (also known as S. s. raveneli Conrad), has a partially sympatric geographic distribution, but differs in several life-history characteristics. DNA sequence variation in mitochondrial cytochrome oxidase I (COI) and in introns at two nuclear calmodulin loci was examined to measure genetic divergence between the two subspecies and to test for population structure among populations of S. s. solidissima. Surfclams were collected from seven localities between 1994 and 2001. Based on both mitochondrial and nuclear DNA variation, the two subspecies of S. solidissima are reciprocally monophyletic, with a net COI divergence of 13.9%, indicating long-term reproductive isolation. The only significant differentiation among populations of S. s. solidissima (based on an AMOVA analysis of COI sequences) was between the Gulf of St. Lawrence and more southerly populations. A long internal branch in the S. s. solidissima genealogy coupled with low haplotype diversity in the northern-most population suggests that populations north and south of Nova Scotia have been isolated from each other in the past, with gene exchange more recently. Populations of S. s. similis from Atlantic and Gulf of Mexico coasts had a net COI divergence of 9.2%. Thus, diversification of Spisula spp. clams in the western North Atlantic involved an early adaptive divergence between coastal and offshore forms, with later barriers to dispersal emerging in the offshore form from north to south and in the coastal form between Atlantic and Gulf of Mexico populations.



Similar content being viewed by others
References
Abbott R (1974) American seashells. Van Nostrand Reinhold, New York
Ambrose WG, Jones DS, Thompson I (1980) Distance from shore and growth rate of the suspension feeding bivalve, Spisula solidissima. Proc Natl Shellfish Assoc 70:207–214
Andrews J (1971) Sea shells of the Texas coast. University of Texas Press, Austin
Avise JC (1992) Molecular population structure and biogeographic history of a regional fauna: a case history with lessons for conservation and biology. Oikos 63:62–76
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge
Avise JC, Ankney CD, Nelson WS (1990) Mitochondrial gene trees and the evolutionary relationship of mallard and black ducks. Evolution 44:1109–1119
Baker AJ, Marshall HD (1997) Mitochondrial control region sequences as tools for understanding evolution. In: Mindell DP (ed) Avian molecular evolution and systematics. Academic, New York, pp 51–82
Bentzen P, Taggart CT, Ruzzante DE, Cook D (1996) Microsatellite polymorphism and the population structure of the Atlantic cod (Gadus morhua) in the northwest Atlantic. Can J Fish Aquat Sci 53:2706–2721
Bernardi G, Sordino P, Powers DA (1993) Concordant mitochondrial and nuclear DNA phylogenies for populations of the teleost fish Fundulus heteroclitus. Proc Natl Acad Sci USA 90:9271–9274
Bernatchez L, Wilson CC (1998) Comparative phylogeography of nearctic and palearctic fishes. Mol Ecol 7:431–452
Bert TM, Arnold WS (1995) An empirical test of predictions of two competing models for the maintenance and fate of hybrid zones: both models are supported in a hard-clam hybrid zone. Evolution 49:276–289
Cerrato RM, Keith DL (1992) Age structure, growth, and morphometric variations in the Atlantic surf clam Spisula solidissima, from estuarine and inshore waters. Mar Biol 114:581–593
Chamberlin JL, Stearns F (1963) A geographic study of the clam, Spisula polynyma (Stimpson). NY Ser Atlas Mar Environ 3:1–12
Chintala MM, Grassle JP (2001) Comparison of recruitment frequency and growth of surfclams, Spisula solidissima (Dillwyn, 1817), in different inner-shelf habitats of New Jersey. J Shellfish Res 20:1177–1186
Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK (2000) Considering evolutionary processes in conservation biology. Trends Ecol Evol 15:290–295
Dahlgren TG, Weinberg JR, Halanych KM (2000) Phylogeography of the ocean quahog (Arctica islandica): influences of paleoclimate on genetic diversity and species range. Mar Biol 137:487–495
David P, Berthou P, Noel P, Jarne P (1997) Patchy recruitment patterns in marine invertebrates: a spatial test of the density-dependent hypothesis in the bivalve Spisula ovalis. Oecologia 111:331–340
Dillon RT, Manzi JJ (1989) Genetics and shell morphology in a hybrid zone between the hard clams Mercenaria mercenaria and Mercenaria campechiensis. Mar Biol 100:217–222
Dillon RT, Manzi JJ (1992) Population genetics of the hard clam, Mercenaria mercenaria, at the northern limit of its range. Can J Fish Aquat Sci 49:2574–2578
Duda TF, Palumbi SR (1999) Developmental shifts and species selection in gastropods. Proc Natl Acad Sci USA 96:10272–10277
Emerson WK, Jacobson MK (1976) Guide to shells. Knopf, New York
Felsenstein J (2004) PHYLIP (Phylogeny Inference Package), ver. 3.6b. University of Washington, Seattle
Folmer O, Black M, Hoeh R, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Fu Y-X (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Gaffney P, Reece KS (2002) Genetic marker development in the eastern oyster. Conference abstract: 6th international conference on shellfish restoration. University of South Carolina, Charleston
Gelfand DH, White TJ (1990) Thermostable DNA polymerases. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, pp 129–135
Giribet G, Wheeler W (2002) On bivalve phylogeny: a high-level analysis of Bivalvia (Mollusca) based on combined morphology and DNA sequence data. Invertebr Biol 121:271–324
Grassly NC, Harvey PH, Holmes EC (1999) Population dynamics of HIV-1 inferred from gene sequences. Genetics 151:427–438
Hare MP (2001) Prospects for nuclear gene phylogeography. Trends Ecol Evol 16:700–706
Hare MP, Avise JC (1996) Molecular genetic analysis of a stepped multilocus cline in the American oyster (Crassostrea virginica). Evolution 50:2305–2315
Hare MP, Avise JC (1998) Population structure in the American oyster as inferred by nuclear gene genealogies. Mol Biol Evol 15:119–128
Hare MP, Palumbi SR, Butman CA (2000) Single-step species identification of bivalve larvae using multiplex polymerase chain reaction. Mar Biol 137:953–961
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Hey J, Nielsen R (2004) Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167:747–760
Hudson RR (1990) Gene genealogies and the coalescent process. Oxf Surv Evol Biol 7:1–44
Hudson RR, Turelli M (2003) Stochasticity overrules the “three-times rule”: genetic drift, genetic draft, and coalescence times for nuclear loci versus mitochondrial DNA. Evolution 57:182–190
Irwin DE (2002) Phylogeographic breaks without geographic barriers to gene flow. Evolution 56:2383–2394
Johannesson H, Stenlid J (2003) Molecular markers reveal genetic isolation and phylogeography of the S and F intersterility groups of the wood-decay fungus Heterobasidion annosum. Mol Phylogenet Evol 29:94–101
Jones DS (1980) Annual cycle of shell growth increment formation in two continental shelf bivalves and its paleoecologic significance. Paleobiology 6:331–340
Kim I, Phillips CJ, Monjeau JA, Birney EC, Noack K, Pumo E, Sikes RS, Dole JA (1998) Habitat islands, genetic diversity, and gene flow in a Patagonian rodent. Mol Ecol 7:667–678
Knowles LL, Maddison WP (2002) Statistical phylogeography. Mol Ecol 11:2623–2635
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245
Lee C (1999) Rapid and repeated invasions of fresh water by the copepod Eurytemora affinis. Evolution 53:1423–1434
Lessios H, Kane J, Robertson D (2003) Phylogeography of the pantropical sea urchin Tripneustes: contrasting patterns of population structure between oceans. Evolution 57:2026–2036
Murawski S, Serchuk F (1989) Mechanized shellfish harvesting and its management: the offshore clam fishery of the eastern United States. In: Caddy J (ed) Marine invertebrate fisheries: their assessment and management. Wiley, New York, pp 479–506
NEFSC (Northeast Fisheries Science Center) (2003) Report of the 37th Northeast Regional Stock Assessment Workshop (37th SAW). C. Atlantic surfclam. Ref. doc. 03-16, NEFSC, Woods Hole, Mass. USA, pp 284–433
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Li W-H (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Neigel JE, Avise JC (1986) Phylogenetic relationships of mitochondrial DNA under various demographic models of speciation. In: Karlin S, Nevo E (eds) Evolutionary processes and theory. Academic, New York, pp 515–534
Nielsen R, Wakeley J (2001) Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158:885–896
Nixon KC, Wheeler QD (1990) An amplification of the phylogenetic species concept. Cladistics 6:211–223
O’Beirn FX, Walker RL, Hurley DH, Moroney DA (1997) Culture of surfclams Spisula solidissima sp., in coastal Georgia: nursury culture. J Shellfish Res 16:157–160
Palumbi SR, Cipriano F, Hare MP (2001) Predicting nuclear gene coalescence from mitochondrial data: the three-times rule. Evolution 55:859–868
Quinn TW (1992) The genetic legacy of mother goose—phylogeographic patterns of lesser snow goose Chen caerulescens caerulescens maternal lineages. Mol Ecol 1:105–117
Reeb CA, Avise JC (1990) A genetic discontinuity in a continuously distributed species: mitochondrial DNA in the America oyster, Crassotrea virginica. Genetics 124:397–406
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Ropes JW (1980) Biological and fisheries data on surf clam, Spisula solidissima (Dillwyn, 1817). US DOC, tech. ser. rep. no. 24, Northeast Fisheries Science Center, Woods Hole, Mass., USA
Rosenberg NA (2003) The shapes of neutral gene genealogies in two species: probabilities of monophyly, paraphyly, and polyphyly in a coalescent model. Evolution 57:1465–1477
Rosenberg NA, Nordborg M (2002) Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Natl Rev Genet 3:380–390
Saunders NC, Kessler LG, Avise JC (1986) Genetic variation and geographic differentiation in mitochondrial DNA of the horseshoe crab, Limulus polyphemus. Genetics 112:613–627
Schizas NV, Coull BC, Chandler GT, Quattro JM (2002) Sympatry of distinct mitochondrial DNA lineages in a copepod inhabiting estuarine creeks in the southeastern USA. Mar Biol 140:585–594
Schneider SD, Excoffier L (1999) Estimation of demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: applications to human mitochondrial DNA. Genetics 152:1079–1089
Schneider SD, Roessli D, Excoffier L (1999) Arlequin ver. 2.0: a software for population genetic data analysis. University of Geneva, Geneva, Switzerland
Schneider-Broussard R, Felder DL, Chlan CA, Neigel JE (1998) Tests of phylogeographic models with nuclear and mitochondrial DNA sequence variation in the stone crabs, Menippe adina and Menippe mercenaria. Evolution 52:1671–1678
Slatkin M, Hudson RR (1991) Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129:555–562
Swofford DL (1998) Phylogenetic analysis using parsimony (PAUP), ver. 4.0. Sinauer, Sunderland, Mass., USA
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Templeton AR (1989) The meaning of species and speciation: a genetic perspective. In: Otte D, Endler JA (eds) Speciation and its consequences. Sinauer, Sunderland, Mass., USA, pp 3–27
Uthicke S, Benzie J (2003) Gene flow and population history in high dispersal marine invertebrates: mitochondrial DNA analysis of Holothuria nobilis (Echinodermata: Holothuroidea) populations from the Indo-Pacific. Mol Ecol 12:2635–2648
Vecchione M, Griffis RB (1996) How many species of surf clams? Oceanography 9:48–49
Wagner E (1984) Growth rate and annual shell structure patterns in a single year class of surfclams Spisula solidissima off Atlantic City, New Jersey. MS thesis, Rutgers University, New Brunswick, N.J., USA
Wakeley J (1995) Distinguishing migration from isolation using the variance of pairwise differences. Theor Popul Biol 49:369–386
Walker RL (1998) Comparative gametogenesis of Spisula solidissima solidissima and Spisula solidissima similis cultured in coastal Georgia. J World Aquacult Soc 29:304–312
Walker RL, Heffernan PB (1994) Age, growth rate, and size of the southern surfclam, Spisula solidissima similis (Say, 1822). J Shellfish Res 13:433–441
Walker RL, O’Beirn FX (1996) Embryonic and larval development of Spisula solidissima similis (Say, 1822) (Bivalvia: Mactridae). Veliger 39:60–64
Walker RL, Hurley DH, Kupfer R (1998) Growth and survival of Atlantic surfclam, Spisula solidissima, larvae and juveniles fed various microalga diets. J Shellfish Res 17:211–214
Wares JP (2002) Community genetics in the northwestern Atlantic intertidal. Mol Ecol 11:1131–1144
Weinberg JR (1998) Density-dependent growth in the Atlantic surfclam, Spisula solidissima, off the coast of the Delmarva Peninsula, USA. Mar Biol 130:621–630
Weinberg JR (1999) Age-structure, recruitment, and adult mortality in populations of the Atlantic surfclam, Spisula solidissima, from 1978 to 1997. Mar Biol 134:113–125
Weinberg JR, Helser TE (1996) Growth of the Atlantic surfclam, Spisula solidissima, from Georges Bank to the Delmarva Peninsula, USA. Mar Biol 126:663–674
Weinberg JR, Dahlgren TG, Trowbridge N, Halanych KM (2003) Genetic differences within and between species of deep-sea crabs (Chaceon) from the North Atlantic Ocean. Biol Bull (Woods Hole) 204:318–326
Williams ST (2000) Species boundaries in the starfish genus Linckia. Mar Biol 136:137–148
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Young AM, Torres C, Mack JE, Cunningham CW (2002) Morphological and genetic evidence for vicariance and refugium in Atlantic and Gulf of Mexico populations of the hermit crab Pagurus longicarpus. Mar Biol 140:1059–1066
Acknowledgements
This study was motivated by preliminary data generated by A.F. Govindarajan with support from a grant to C.A. Butman for an unrelated study. We thank C. Salas, P. Gaffney, J. Grassle, C. Gregg, R. Morneau, F. O’Beirn, and R. Walker for providing samples or collection data. We are also grateful for collection efforts by the scientists and crew of NOAA’s R.V. “Delaware II”. M.P.H. is grateful for support from the Hrdy Fellowship in Conservation Biology at Harvard University, where he was hosted by S. Palumbi. Further support to M.P.H. was obtained from the General Research Board at the University of Maryland.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.P. Grassle, New Brunswick
Rights and permissions
About this article
Cite this article
Hare, M.P., Weinberg, J.R. Phylogeography of surfclams, Spisula solidissima, in the western North Atlantic based on mitochondrial and nuclear DNA sequences. Marine Biology 146, 707–716 (2005). https://doi.org/10.1007/s00227-004-1471-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-004-1471-y