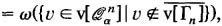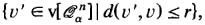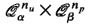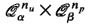Abstract
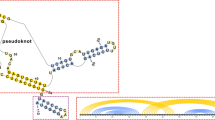
Random graph theory is used to model and analyse the relationship between sequences and secondary structures of RNA molecules, which are understood as mappings from sequence space into shape space. These maps are non-invertible since there are always many orders of magnitude more sequences than structures. Sequences folding into identical structures formneutral networks. A neutral network is embedded in the set of sequences that arecompatible with the given structure. Networks are modeled as graphs and constructed by random choice of vertices from the space of compatible sequences. The theory characterizes neutral networks by the mean fraction of neutral neighbors (λ). The networks are connected and percolate sequence space if the fraction of neutral nearest neighbors exceeds a threshold value (λ>λ*). Below threshold (λ<λ*), the networks are partitioned into a largest “giant” component and several smaller components. Structure are classified as “common” or “rare” according to the sizes of their pre-images, i.e. according to the fractions of sequences folding into them. The neutral networks of any pair of two different common structures almost touch each other, and, as expressed by the conjecture ofshape space covering sequences folding into almost all common structures, can be found in a small ball of an arbitrary location in sequence space. The results from random graph theory are compared to data obtained by folding large samples of RNA sequences. Differences are explained in terms of specific features of RNA molecular structures.
Similar content being viewed by others
Abbreviations
- v[G]:
-
Vertex set of graphG
- e[G]:
-
Edge set of graphG
- ω(X):
-
Cardinality ofX as a set
- δ v :
-
Vertex degree in a corresponding graphG
- Q n α :
-
Generalized hypercube
- \(\hat X\) :
-
X is a random variable
- E[\(\hat X\)]:
-
Expectation value of the random variable\(\hat X\)
- V[\(\hat X\)]:
-
Variance of\(\hat X\)
- E[\(\hat X\)] r :
-
rth factorial moment of\(\hat X\)
- μ n, λ,μ n :
-
Measure\(\mu _n (\Gamma _n )\mathop = \limits^{def} \lambda ^{\omega (v[\Gamma ])} (1 - \lambda )^{\omega (v[H]) - \omega (v[\Gamma ])} \)
- Ω n :
-
Probability space ({Γ n },μ n, λ
- \(\hat X_{n,k} \) :
-
Number of vertices in a random graph Γ n having degreek
- \(\hat I_n (\Gamma _n )\) :
-
=ω({v∈v[Γ n ]|∂{v}∩v[Γ n ]=∅}), i.e., the number of isolated vertices in a random graph Γ n
- \(\hat Z_n (\Gamma _n )\) :
-
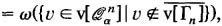
i.e. the number of vertices inQ n α that are at least of distance 2 w.r.t. a random graph Γ n
- \(M_{n,k}^{\upsilon ,\upsilon '} (\Gamma _n )\) :
-
Set of paths {π(υ1)|π(υ1)∈Π(Γ n )}
- \(\hat Y_{n,k}^{\upsilon ,\upsilon '} \) :
-
\( = \omega (M_{n,k}^{\upsilon ,\upsilon '} (\Gamma _n ))\) and 0 otherwise
- \(\hat \Lambda _{n,k} \) :
-
Random variable that is 1 if all pairs υ,υ′∈v[Γ n ] withd(v, v′)<k occur in a path ofd(υ,υ′)<k and 0 otherwise
- ∂ G V :
-
Set of adjacent vertices w.r.t. a vertex setV⊂v[G] in a graphG
- \(\bar V\) :
-
v[V]∪∂V, i.e. the closure ofV
- ℬ r (v):
-
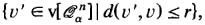
the “ball” with radiusr and centerv
- n :
-
Chain length
- n u ,n p :
-
Number of unpaired and paired based of a certain secondary structure
- γ n :
-
(α−1)n, i.e. the vertex degrees ofQ n α
- s :
-
RNA secondary structure inn vertices
- Π(s :
-
\(\mathop = \limits^{def} \left\{ {\left[ {i,k} \right]|a_{i,k} = 1,k \ne i - 1,i + 1} \right\}\) i.e. the set of contacts of the secondary structures
- L n :
-
Shape space, in particular, the space of RNA secondary structures inn vertices
- C[s]:
-
Graph of compatible sequences with respect tos
- C[s]:
-
v[C[s]], the set of compatible sequences
- S n :
-
Permutation group ofn letters
- D m :
-
Dihedral group of order2m
- Φ Γ x :
-
G 1 ×G 2{y∈v[G 2]⋎(x, y)∈v[⩾s]}], the fiber inx
- Φ Γ y :
-
G 1 ×G 2[{x∈v[G 1]⋎(x, y)∈v[⩽s]}], the fiber iny
- Γ A n [s]:
-
Random induced subgraph of
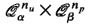
according to model A
- Γ B n [s]:
-
Random induced subgraph of
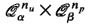
according to model B
- dist(Γ1, Γ2):
-
Minimum Hamming distance between the graph Γ1 and Γ2 considered as subgraph ofQ n α
References
Ajtai, M., J. Komlós and E. Szemerédi. 1982. Largest random component of ak-cube.Combinatorica,2, 1–7.
Bollobás, B. 1985.Random Graphs. London: Academic Press.
Bonhoeffer, S. and P. F. Stadler. 1993. Error threshold on complex fitness landscapes.J. Theor. Biol. 164, 359–372.
Buckley, F. and F. Harrary. 1990.Distances in Graphas Reading, MA: Addison-Wesley.
Derrida, B. and L. Peliti. 1991. Evolution in a flat fitness landscape.Bull. Math. Biol. 53, 355–382.
Eigen, M. 1971. Self-organization of matter and the evolution of biological macromolecules.Naturwissenschaften 58, 465–523.
Eigen, M., J. McCaskill and P. Schuster. 1989. The molecular quasispecies.Adv. Chem. Phys. 75, 149–263.
Eigen, M. and P. Schuster. 1977. The hypercycle. A principle of natural self-organization. Part A: emergence of the hypercycle.Naturwissenschaften 64, 541–565.
Fontana, W. T. Griesmacher, W. Schnabl, P. F. Stadler and P. Schuster. 1991. Statistics of landscapes based on free energies, replication and degradation rate constants of RNA secondary structures.Mh. Chem. 122, 795–819.
Fontana, W., W. Schnabl and P. Schuster. 1989. Physical aspects of evolutionary optimization and adaptation.Phys. Rev. A 40, 3301–3321.
Fontana, W. and P. Schuster. 1987. A computer model of evolutionary optimization.Biophys. Chem. 26, 123–147.
Fontana, W., D. A. M. Konings, P. F. Stadler and P. Schuster. 1993a. Statistics of RNA secondary structures.Biopolymers 33, 1389–1404.
Fontana, W., P. F. Stadler, E. G. Bornberg-Bauer, T. Griesmacher, I. L. Hofacker, M. Tacker, P. Tarazona, E. D. Weinberger and P. Schuster. 1993b. RNA folding and combinatory landscapes.Phys. Rev. E 47, 2083–2099.
Freier, S., R. Kierzek, J. Jaeger, N. Sugimoto, M. Caruthers, T. Neilson and D. Turner. 1986. Improved free-energy parameters for predictions of RNA duplex stability.Proc. Natl. Acad. Sci. USA 83, 9373–9377.
Grüner, W., R. Giegerich, D. Strothmann, C. Reidys, J. Weber, I. L. Hofacker, P. F. Stadler and P. Schuster. 1996a. Analysis of RNA sequence structure maps by exhaustive enumeration. I. Neutral networks.Mh. Chem. 127, 355–374.
Grüner, W., R. Giegerich, D. Strothmann, C. Reidys, J. Weber, I. L. Hofacker, P. F. Stadler and P. Schuster. 1996b. Analysis of RNA sequence structure maps by exhaustive enumeration. II. Structure of neutral networks and shape space covering.Mh. Chem. 127, 375–389.
Hamming, R. W. 1950. Error detecting and error correcting codes.Bell Syst. Tech. J 29, 147–160.
Harper, L. 1966. Minimal numberings and isoperimetric problem on cubes.Theory of Graphs, International Symposium. Rome.
Hofacker, I. L. 1994.A Statistical Characterization of the Sequence to Structure Mapping in RNA. Ph.D. thesis, Universität Wien.
Hofacker, I. L., W. Fontana, P. F. Stadler, M. Bonhoeffer, M. Tacker and P. Schuster. 1994. Fast folding and comparison of RNA secondary structures.Mh. Chem. 125, 167–188.
Hofacker, I. L., P. Schuster and P. F. Stadler. 1996. Combinatorics of RNA secondary structures. Preprint.
Hogeweg, P. and B. Hesper. 1984. Energy directed folding of RNA sequences.Nucleic Acids Res. 12, 67–74.
Huynen, M. A., P. F. Stadler and W. Fontana. 1996. Smoothness within ruggedness: the role of neutrality in adaptation.Proc. Natl. Acad. Sci. USA 93, 397–401.
Kesten, H. 1982.Percolation Theory for Mathematics. Boston, MA: Birkhäuser.
Kimura, M. 1983.The Neutral Theory of Molecular Evolution Cambridge: Cambridge University Press.
Konings, D. and P. Hogeweg. 1989. Pattern analysis of RNA secondarys structure. Similarity and consensus of minimal-energy folding.J. Mol. Biol. 207, 597–614.
Li, H., R. Helling, C. Tang and N. Wingreen 1996. Emergence of preferred structures in a simple model of protein folding.Science 273, 666–669.
Martinez, H. 1984. An RNA folding rule.Nucleic Acids Res. 12, 323–335.
McCaskill, J. 1990. The equilibrium partition function and base pair bindings probabilities for RNA secondary structure.Biopolymers 29, 1105–1119.
Nowak, M. and P. Schuster. 1989. Error thresholds of replication in finite populations. Mutation frequencies and the onset of Muller's ratchet.J. Theor. Biol. 137, 375–395.
Nussinov, R., G. Piecznik, J. Griggs and D. Kleitman. 1978. Algorithms for loop matching.SIAM J. Appl. Math. 35, 68–82.
Reidys, C. 1995.Neutral Networks of RNA Secondary Structures. Ph.d. thesis, Friedrich-Schiller-Universität Jena.
Reidys, C., C. V. Forst and P. Schuster. 1996. Replication on neutral networks of RNA secondary structures. Preprint.
Riordan, J. 1978.An Introduction to Combinatorial Analysis. Princetons, NJ: Princeton University Press.
Salser, W. 1977. Globin messenger RNA sequences—analysis of base-pairing and evolutionary implications.Cold Spring Harbor Symp. Quant. Biol. 42, 985.
Sankoff, D., G. Leduc, N. Antoine, B. Pacquin, B. F. Lang and R. Cedergren. 1992. Gene comparisons for phylogenetic inference: evolution of the mitochondrial genome.Proc. Natl. Acad. Sci. USA 89, 6575–6579.
Schuster, P. 1995a. Artificial life and molecular evolutionary biology. InAdvances in Artificial Life. F. Morán, A. Moreno, J. J. Morelo and P. Chacón (Eds).Lecture Notes in Artificial Intelligence, Vol. 929, pp. 3–19, Berlin: Springer-Verlag.
Schuster, P. 1995b. How to search for RNA structures. Theoretical concepts in evolutionary biotechnology.J. Biotechnol. 41, 239–257.
Schuster, P., W. Fontana, P. F. Stadler and K. L. Hofacker. 1994. From sequences to shapes and back: a case study in RNA secondary structures.Proc. Roy. Soc. (London) B 255, 279–284.
Schuster, P. and P. F. Stadler. 1994. Landscapes: complex optimization problems and biopolymer structures.Computers Chem. 18, 295–314.
Serre, J.-P. 1977.Linear Representations of Finite Groups. Berlin: Springer.
Shapiro, B. A. 1988. An algorithm for comparing multiple RNA secondary structures.CABIOS 4, 381–393.
Shapiro, B. A. and K. Zhang. 1990. Comparing multiple RNA secondary structures using tree comparisons.CABIOS 6, 309–318.
Stauffer, D. 1985.Introduction to Percolation Theory. London: Taylor and Francis.
Tacker, M., W. Fontana, P. F. Stadler and P. Schuster. 1994. Statistics of RNA melting kinetics.Eur. Biophys. J. 23, 29–38.
Tacker, M., P. F. Stadler, E. G. Bornberg-Bauer, K. L. Hofacker and P. Schuster. 1996. Algorithm independent properties of RNA secondary structure predictions.Eur. Biophys. J. 25, 115–130.
Turner, D. H., N. Sugimoto and S. Freier. 1988. RNA structure prediction.Annual Review of Biophysics and Biophysical Chemistry 17, 167–192.
Waterman, M. S. 1978. Secondary structure of single-stranded nucleic acids.Adv in Math. 1, 167–212.
Zuker, M. and D. Sankoff. 1984. RNA secondary structures and their prediction.Bull. Math. Biol. 46, 591–621.
Zuker, M. and P. Stiegler. 1981. Optimal computer folding of larger RNA sequences using thermodynamics and auxiliary information.Nucleic Acids Res. 9, 133–148.
Author information
Authors and Affiliations
Additional information
Deicated to professor Manfred Eigen
Rights and permissions
About this article
Cite this article
Reidys, C., Stadler, P.F. & Schuster, P. Generic properties of combinatory maps: Neutral networks of RNA secondary structures. Bltn Mathcal Biology 59, 339–397 (1997). https://doi.org/10.1007/BF02462007
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02462007