Abstract
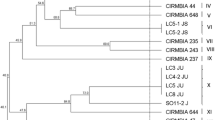
The aims of this work were to assess the levels of Lactococcus lactis (L. lactis) in ewe’s milk produced in three Ossau-Iraty cheese sub-areas and to investigate the genotypic and technological diversity of isolated wild strains of L. lactis in order to assess their suitability for use as components of starter formulations. Thirty-two milk samples from 32 farms were collected. Strains of L. lactis were identified and quantified using a combination of species and subspecies-specific polymerase chain reaction (PCR) and PCR amplification of repetitive bacterial DNA elements (Rep-PCR). The genotypic and technological diversity of the indigenous strains was compared to that of 12 commercial strains. L. lactis was detected in milk samples from only 20 farms. The levels detected were below 4 log10 cfu.mL−1 in 75% of the milks. L. lactis subsp. lactis dominated in 66% of the samples. Forty-three genotypic profiles of wild L. lactis strains were detected and showed greater diversity than those of the commercial strains. Milks containing L. lactis contained one to four distinct strains. With the exception of two strains, each strain was found in milk from only one farm. The Prt+ strains were the most acidifying. Sensitivity to phages collected from wheys differed widely between the commercial (60%) and indigenous strains (5%). Wild strains of L. lactis displayed a wide genotypic and technological diversity. Genotypic diversity seemed to be linked to the farm of origin. This study addresses questions regarding the environmental factors which influence such natural diversity. A deeper knowledge of the strain-dependent technological properties would be useful in selecting strains for use in starter blends.
摘要:
本文目的是评价Ossau-Iraty原产地名号保护 (PDO) 干酪生产地区生鲜羊乳乳酸乳球菌的含量以及分离的野生乳酸乳球菌的基因型和多样性, 以及这些乳酸菌作为发酵剂的可行性. 32个乳样分别来自32个农场. 采用物种和亚种特异性聚合酶链反应(PCR)和重复细菌DNA元素PCR扩增相结合的方法来分析和鉴定乳酸乳球菌. 将分离菌株的基因型和技术特性与12株商业菌株进行了比较, 仅从20个农场的羊乳样本中检测到乳酸乳球菌. 75%的乳样中乳酸乳球菌的含量低于4 log cfu mL-1, 乳酸乳球菌乳酸亚种在样品中占66%. 野生乳酸乳球菌中检测出43种基因型, 与商业菌株相比, 野生乳酸乳球菌更具有多样性. 乳酸乳球菌中含有1∼4株特殊的菌株. 除2株菌外, 每个农场的羊奶样品中都能够检测到不同于其他农场的特有菌株. 从乳清中收集的噬菌体与商业菌株和本土菌株比较存在较大的差异. 蛋白酶阳性 (Prt +) 菌株的酸化能力最强. 野生乳酸乳球菌显示了较广的基因型和多样性. 基因多样性似乎与农场有关. 本研究进一步说明了环境因素是影响菌株固有多样性的主要因素. 对菌株技术特性的深入研究将有助于混合发酵剂菌株的筛选.



Similar content being viewed by others
References
Ayad EHE, Verheul A, Wouters JTM, Smit G (2000) Application of wild starter cultures for flavor development in pilot plant cheese making. Int Dairy J 10:169–179
Ayad EHE, Nashat S, El-adek N, Metwaly M, El-Soda M (2004) Selection of wild lactic acid bacteria isolated from traditional Egyptian dairy products according to production and technological criteria. Food Microbiol 21:715–725
Barakat RK, Griffiths MW, Harris LJ (2000) Isolation and characterization of Carnobacterium, Lactococcus and Enterococcus spp. from cooked, modified atmosphere packaged, refrigerated, poultry meat. Int J Food Microbiol 62:83–94
Beimfohr C, Ludwig W, Schleifer KH (1997) Rapid genotypic differentiation of Lactococcus lactis subspecies and biovar. Syst Appl Microbiol 20:216–221
Berthier F, Beuvier E, Dasen A, Grappin R (2001) Origin and diversity of mesophilic lactobacilli in Comté cheese, as revealed by PCR with repetitive and species-specific primers. Int Dairy J 11:293–295
Bruinenberg PG, Vos P, de Vos WM (1992) Proteinase overproduction in Lactococcus lactis strains: regulation and effect on growth and acidification in milk. Appl Env Microbiol 58:78–84
Callon C, Millet L, Montel MC (2004) Diversity of lactic acid bacteria isolated from AOC Salers cheese. J Dairy Res 71:231–244
Casalta E, Montel MC (2008) Safety assessment of dairy microorganisms: the Lactococcus genus. Int J Food Microbiol 126:271–273
Casalta E, Noël Y, Le Bars D, Carré C, Achilleos C, Maroselli MX (2001) Caractérisation du fromage Bastelicaccia. Lait 81:529–546
Casalta E, Vassal Y, Desmazeaud MJ, Casabianca F (1995) Comparaison de l’activité acidifante des souches de Lactococcus lactis isolées de lait et de fromages de Corse. Leben Wissen Technol 28:291–299
Corroler D, Mangin I, Desmasures N, Guéguen M (1998) An ecological study of lactococci isolated from raw milk in the Camembert cheese Registered Designation of Origin area. Appl Env Microbiol 64:4729–4735
de La Plaza M, Rodriguez A, Fernandez de Palencia P, Martinez-Cuseta MC, Pelaez C, Requena T (2006) Discrepancies between the phenotypic and genotypic characterization of Lactococcus lactis cheese isolates. Lett Appl Microbiol 43:637–644
de Godoy Oriani MR, Yokoya F (2004) Lactococcus bacteriophages isolated from whey and their effects on commercials lactic starters. Braz Arch Biol Technol 47:559–568
Depouilly A, Dufrene F, Beuvier E, Berthier F (2004) Genotypic characterization of the dynamics of the lactic acid bacterial population of Comté cheese. Lait 84:155–167
Desmasures N, Mangin I, Corroler D, Guéguen M (1998) Characterization of lactococci isolated from milk produced in the Camembert region of Normandy. J Appl Microbiol 85:999–1005
Fernández E, Alegría A, Delgado S, Martín MC, Mayo B (2011) Comparative phenotypic and molecular genetic profiling of wild Lactococcus lactis subsp. lactis strains of the L. lactis subsp. lactis and L. lactis subsp. cremoris genotypes isolated from starter-free cheeses made of raw milk. Appl Env Microbiol 77:5324–5335
Feutry F, Oneca M, Berthier F, Torre P (2011) Biodiversity of lactic acid bacteria in raw ewe’s milk PDO Ossau-Iraty cheeses made with different starters. Food Microbiol 29:33–42
Franciosi E, Settani L, Cavazza A, Poznanski E (2009) Biodiversity and technological potential of wild lactic acid bacteria from raw cow’s milk. Int Dairy J 19:3–11
Gaya P, Babin M, Medina M, Nuñez M (1999) Diversity among lactococci isolated from ewe’s raw milk cheese. J Appl Microbiol 87:849–855
IDF (International Dairy Federation) (1996) Preparation of samples and dilutions for microbiological examination. Standard 122C. Brussels. Int Dairy Fed.
Jeanson S, Berthier F, Grappin R, Beuvier E (2003) Heat resistance of wild Lactococcus lactis strains under a thermal gradient of cooked cheese, in milk and in mini-cheeses. Lait 83:1–16
Klijn N, Weerkamp AH, de Vos WM (1995) Detection and characterization of lactose-utilizing Lactococcus spp. in natural ecosystems. Appl Env Microbiol 61:788–792
Labrie S, Moineau S (2000) Multiplex PCR for detection and identifiaction of Lactococcal bacteriophages. Appl Env Microbiol 66:987–994
Lafarge V, Ogier JC, Girard V, Maladen V, Leveau JY, Gruss A, Delacroix-Buchet A (2004) Raw cow milk bacterial population shifts attributable to refrigeration. App Env Microbiol 70:5644–5660
Mannu L, Paba A, Pes M, Scintu MF (2000) Genotypic and phenotypic heterogeneity among lactococci isolated from traditional Pecorino Sardo cheese. J Appl Microbiol 89:191–197
Martín MC, Ladero V, Alvarez MA (2006) PCR identification of lysogenic Lactococcus lactis strains. Journal of Consumer Protection and Food Safety 1:121–124
Medina R, Katz M, Gonzalez S, Oliver G (2001) Characterization of the lactic acid bacteria in ewe’s milk and cheese from Northwest Argentina. J Food Prot 64:559–563
Nieto-Arribas P, Seseña S, Poveda JM, Palop L, Cabezas L (2009) Genotypic and technological characterization of Lactococcus lactis isolates involved in processing of artisanal Manchego cheese. J Appl Microbiol 107:1505–1517
Oneca M, Irigoyen A, Ortigosa M, Torre P (2003) PCR and RAPD identification of L. plantarum strains isolated from ovine milk and cheese. Geographical distribution of strains. FEMS Microbiol Lett 227:271–277
Pérez-Elortondo FJ, Albisu M, Barcina Y (1993) Changes in the microflora of Idiazábal cheese with the addition of commercial lactic starters. Aust J Dairy Technol 48:10–14
Reiter B, Kirikova M (1976) The isolation of a lysogenic strains from a multiple strains starter culture. J Soc Dairy Technol 29:221–225
Sánchez MM, Delgado T, Alonso L, Mayo B (2000) Phenotypic and genetic characterization of a selected set of Lactococcus lactis strains isolated from a starter-free farmhouse cheese. Food Microbiol 17:449–460
Taïbi A, Dabour N, Lamoureux M, Roy D, La Pointe G (2010) Evaluation of the genetic polymorphism among Lactococcus lactis subsp. cremoris strains using comparative genomic hybridization and multilocus sequence analysis. Int J Food Microbiol 144:20–28
Terzaghi BE, Sandine WE (1975) Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol 29:807–813
Versalovic J, Koeuth T, Lupski JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 24:6823–6831
Ward LJH, Heap HA, Kelly WJ (2004) Characterization of closely related lactococcal starter strains which show differing patterns of bacteriophage sensitivity. J Appl Microbiol 96:144–148
Acknowledgments
This work was financially supported by the Aquitaine Regional Council, the Pyrénées Atlantiques Council, ONILAIT (Paris, France), the French State (FNADT), and the European Commission (Leader II and Interreg IIIA programs). The authors would like to thank T. Janzen (Christian Hansen, Hoersholm, Denmark), E. Suárez, and C. Madera (Oviedo University, Spain) for supplying the phages and the producers from whom the milk samples were taken.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Feutry, F., Torre, P., Arana, I. et al. Lactococcus lactis strains from raw ewe’s milk samples from the PDO Ossau-Iraty cheese area: levels, genotypic and technological diversity. Dairy Sci. & Technol. 92, 655–670 (2012). https://doi.org/10.1007/s13594-012-0084-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13594-012-0084-3