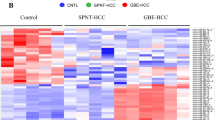
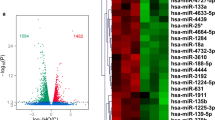
Abstract
Toxicology studies assessing the risk of environmental toxicants in humans frequently use in vitro systems in combination with transcriptomics to characterize toxic responses. Thus far, changes have mostly been investigated at the mRNA level. Recently, microRNAs (miRNAs) have attracted attention because they are powerful negative regulators of mRNA levels and thus may be responsible for the modulation of important mRNA networks implicated in toxicity. This study aimed to identify possible miRNA-mRNA networks as novel interactions at the gene expression level after exposure to environmental toxicants. Benzo[k]fluoranthene (BF), a polycyclic aromatic hydrocarbon that is ubiquitously distributed throughout the environment, was used. We analyzed mRNA and miRNA profiles in HepG2 cells, a human liver cell line, using a human oligonucleotide chip. Changes in miRNA expression in response to BF, in combination with multiple alterations of mRNA levels, were observed. Many of the altered mRNAs were targets of altered miRNAs. Using gene ontology (GO) and KEGG pathway analysis, we determined the relevance of such miRNA deregulation to carcinogenicity. This revealed five miRNAs that appear to participate in specific BFresponsive pathways relevant to genotoxicity and carcinogenicity, such as DNA damage repair, apoptosis, cancer, VEGF signaling, and Jak-STAT signaling. Our results indicate that miR-146a, miR-365, let-7f, miR-199b-5p, and miR-30c-1* are novel players in the BF response. Therefore, this study demonstrates the added value of an integrated miRNA-mRNA approach for identification of the molecular mechanisms induced by BF in an in vitro human model.
Similar content being viewed by others
References
Rana, T. M. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 8, 23–36 (2007).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005).
Volinia, S. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 103, 2257–2261 (2006).
Taylor, E. L. & Gant, T. W. Emerging fundamental roles for non-coding RNA species in toxicology. Toxicology 246, 34–39 (2008).
Brueckner, B. et al. The human let-7a-3 locus contains an epigenetically regulated MicroRNA gene with oncogenic function. Cancer Res. 67, 1419–1423 (2007).
Johnson, S., Lin, S. & Slack, F. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 259, 364–379 (2003).
Meng, F. et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 130, 2113–2129 (2006).
Pogribny, I. P. et al. Induction of microRNAome deregulation in rat liver by long-term tamoxifen exposure. Mutat. Res. 619, 30–37 (2007).
Sathyan, P., Golden, H. B. & Miranda, R. C. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J. Neurosci. 27, 8546–8557 (2007).
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 (2004).
Pothof, J. et al. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 28, 2090–2099 (2009).
Kumar, M. S., Lu, J., Mercer, K. L., Golub, T. R. & Jacks, T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39, 673–677 (2007).
Guo, H., Ingolia, N. T., Weissman, J. S. & Bartel, D. P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010).
Sun, B. K. & Tsao, H. Small RNAs in development and disease. J. Am. Acad. Dermatol. 59, 725–737 (2008).
Wang, Y., Liang, Y. & Lu, Q. MicroRNA epigenetic alterations: Predicting biomarkers and therapeutic targets in human diseases. Clin. Genet. 74, 307–315 (2008).
Lynam-Lennon, N., Maher, S. G. & Reynolds, J. V. The roles of micro-RNA in cancer and apoptosis. Biol. Rev. Camb. Philos. Soc. 84, 55–71 (2009).
Liu, H. & Kohane, I. S. Tissue and process specific microRNA-mRNA co-expression in mammalian development and malignancy. PLoS One 4, e5436 (2009).
Lafferty-Whyte, K., Cairney, C. J., Jamieson, N. B., Oien, K. A. & Keith, W. N. Pathway analysis of sene-scence-associated miRNA targets reveals common processes to different senescence induction mechanisms. Biochim. Biophys. Acta. 1792, 341–352 (2009).
Fukushima, T., Hamada, Y., Yamada, H. & Horii, I. Changes of micro-RNA expression in rat liver treated by acetaminophen or carbon tetrachloride-regulating role of micro-RNA for RNA expression. J. Toxicol. Sci. 32, 401–409 (2007).
Wang, K. et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 106, 4402–4407 (2009).
Agency for Toxic Substances and Disease Registry. Toxicological Profile for Polycyclic Aromatic Hydrocarbons (PAHs). Atlanta, GA: ATSDR (1995).
Mastrangelo, G., Fadda, E. & Marzia, V. Polycyclic aromatic hydrocarbons and cancer in man. Environ. Health Perspect. 104, 1166–1170 (1996).
Song, M. K. et al. Gene expression analysis identifies DNA damage-related markers of benzo[a]pyrene exposure in HepG2 human hepatocytes. Toxicol. Environ. Health. Sci. 4, 19–29 (2012).
Izzotti, A. et al. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 23, 806–812 (2009).
Schembri, F. et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl. Acad. Sci. USA 106, 2319–2324 (2009).
Bolleyn, J. et al. Effect of Trichostatin A on miRNA expression in cultures of primary rat hepatocytes. Toxicol. in Vitro 25, 1173–1182 (2011).
Yauk, C. L., Jackson, K., Malowany, M. & Williams, A. Lack of change in microRNA expression in adult mouse liver following treatment with benzo(a)pyrene despite robust mRNA transcriptional response. Mutat. Res. 722, 131–139 (2010).
Halappanavar, S. et al. Pulmonary gene and microRNA expression changes in mice exposed to benzo(a)pyrene by oral gavage. Toxicology 285, 133–141 (2011).
Duan, H., Jiang, Y., Zhang, H. & Wu, Y. MiR-320 and miR-494 affect cell cycles of primary murine bronchial epithelial cells exposed to benzo[a]pyrene. Toxicol. in Vitro 24, 928–935 (2010).
Lema, C. & Cunningham, M. J. MicroRNAs and their implications in toxicological research. Toxicol. Lett. 198, 100–105 (2010).
Yokoi, T. & Nakajima, M. Toxicological implications of modulation of gene expression by microRNAs. Toxicol. Sci. 123, 1–14 (2011).
Audebert, M. et al. Use of the gammaH2AX assay for assessing the genotoxicity of polycyclic aromatic hydrocarbons in human cell lines. Toxicol. Lett. 199, 182–192 (2010).
Song, M. K., Kim, Y. J., Song, M., Choi, H. S. & Ryu, J. C. Dose-response functional gene analysis by exposure to 3 different polycyclic aromatic hydrocarbons in human hepatocytes. Mol. Cell. Toxicol. 7, 221–232 (2011).
Yokoi, T. & Nakajima, M. Toxicological implications of modulation of gene expression by microRNAs. Toxicol. Sci. 123, 1–14 (2011).
Sonkoly, E. & Pivarcsi, A. MicroRNAs in inflammation and response to injuries induced by environmental pollution. Mutat. Res. 17, 46–53 (2011).
Moffat, I. D. et al. microRNAs in adult rodent liver are refractory to dioxin treatment. Toxicol. Sci. 99, 470–487 (2007).
Malik, A. I., Williams, A., Lemieux, C. L., White, P. A. & Yauk, C. L. Hepatic mRNA, microRNA, and miR-34a-Target responses in mice after 28 days exposure to doses of benzo(a)pyrene that elicit DNA damage and mutation. Environ. Mol. Mutagen. 53, 10–21 (2012).
Farh, K. K. et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310, 1817–1821 (2005).
Favaro, E. et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One 5, e10345 (2010).
Sethi, J. K. & Vidal-Puig, A. Wnt signalling and the control of cellular metabolism. Biochem. J. 427, 1–17 (2010).
Hutcheson, J. et al. Combined deficiency of proapoptotic regulators Bim and Fas results in the early onset of systemic autoimmunity. Immunity 28, 206–217 (2008).
Zhang, L. et al. Integrative genomic analysis of phosphatidylinositol 3′-kinase family identifies PIK3R3 as a potential therapeutic target in epithelial ovarian cancer. Clin. Cancer Res. 13, 5314–5321 (2007).
Wang, H. Q. et al. Deregulated miR-155 promotes Fasmediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J. Pathol. 225, 32–42 (2011).
Crowder, R. J. et al. PIK3CA and PIK3CB inhibition produce synthetic lethality when combined with estrogen deprivation in estrogen receptor-positive breast cancer. Cancer Res. 69, 3955–3962 (2009).
Qin, B. et al. MicroRNAs expression in ox-LDL treated HUVECs: MiR-365 moudlates apoptosis and Bcl-2 expression. Biochem. Biophys. Res. Commun. 410, 127–133 (2011).
Li, J. et al. miR-146a, an IL-1β responsive miRNA, induces vascular endothelial growth factor and chondrocyte apoptosis by targeting Smad4. Arthritis Res. Ther. 14, R75 (2012).
Dang, C. V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19, 1–11 (1999).
Cazzalini, O., Scovassi, A. I., Savio, M., Stivala, L. A. & Prosperi, E. Multiple roles of the cell cycle inhibitor p21 (CDKN1A) in the DNA damage response. Mutat. Res. 704, 12–20 (2010).
Sujobert, P. et al. Essential role for the p110d isoform in phosphoinositide 3-kinase activation and cell proliferation in acute myeloid leukemia. Blood 106, 1063–1066 (2005).
Mosmann, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 (1983).
Park, H. W., Kim, S. J. & Oh, M. J. Gene expression patterns of environmental chemicals in human cell lines using HazChem human array. BioChip J. 3, 65–70 (2009).
Yim, W. C. et al. Identification of novel 17β-estradiol (E2) target genes using crossexperiment gene expression datasets. Toxicol. Environ. Health. Sci. 2, 25–38 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 289–300 (1995).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Song, MK., Song, M., Choi, HS. et al. Benzo[k]fluoranthene-induced changes in miRNA-mRNA interactions in human hepatocytes. Toxicol. Environ. Health Sci. 4, 143–153 (2012). https://doi.org/10.1007/s13530-012-0129-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13530-012-0129-2