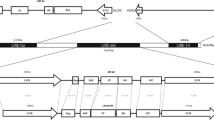
Abstract
Transposable elements are key players in eukaryotic genomic diversity. Due to their high abundance, great heterogeneity, and replicative transposition, long terminal repeat (LTR) retrotransposons are well suited as molecular markers for breeding and biodiversity studies, whereas their utilities in non-model organisms have been hindered by the lack of prior sequence knowledge. In this study, a putative complete (5362 bp) LTR retrotransposon was isolated and characterized in the mangrove species Kandelia candel (named KCRE1), and its transcription, insertional polymorphisms and copy number variations were also investigated. KCRE1 has all the features of a typical copia retroelement, and its transcription initiation and termination sites were identified by 5′ and 3′ rapid amplification of cDNA ends (RACE), respectively. KCRE1 exhibits high sequence similarity with the tomato retroelement Rider and is constitutively expressed in the leaf, root, flower, and hypocotyl tissues of K. candel. Based on KCRE1, sequence-specific amplification polymorphism (SSAP) markers were developed to explore genetic diversity across 30 individuals from three natural populations of K. candel in China. Six primer combinations yielded a total of 204 SSAP bands with an averaged percentage of polymorphic loci of 55.37 and 41.35 % at the species and population levels, respectively. Each individual had a distinct SSAP phenotype, and 14–23 unique bands were observed for each population. Accordingly, KCRE1 was highly abundant in the genome of K. candel and showed considerable copy number variation among the three populations. In conclusion, KCRE1 is the first transcriptionally active retrotransposon reported in K. candel, providing a useful tool for the elucidation of untapped genetic diversity in mangrove genomes.




Similar content being viewed by others
References
Bardil A, Tayale A, Parisod C (2015) Evolutionary dynamics of retrotransposons following autopolyploidy in the Buckler Mustard species complex. Plant J 82:621–631
Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42:251–269
Beulé T, Agbessi MD, Dussert S, Jaligot E, Guyot R (2015) Genome-wide analysis of LTR-retrotransposons in oil palm. BMC Genom 16:795. doi:10.1186/s12864-015-2023-1
Biswas MK, Chai L, Amar MH (2011) Comparative analysis of genetic diversity in Citrus germplasm collection using AFLP, SSAP, SAMPL and SSR markers. Sci Hortic 129:798–803
Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13:3261–3273
Cavrak VV, Lettner N, Jamge S, Kosarewicz A, Bayer LM, Mittelsten Scheid O (2014) How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet 10:e1004115. doi:10.1371/journal.pgen.1004115
Chen X-Y (1999) Population genetics and evolution of mangroves. In: Zu YG, Sun M, Kang L (eds) The application, method and theory of molecular ecology. China Higher Education Press, Beijing, pp 206–216 (in Chinese with English abstract)
Cheng X, Zhang D, Cheng Z, Keller B, Ling HQ (2009) A new family of Ty1-copia-like retrotransposons originated in the tomato genome by a recent horizontal transfer event. Genetics 181:1183–1193
Chesnay C, Kumar A, Pearce SR (2007) Genetic diversity of SIRE-1 retroelements in annual and perennial Glycine species revealed using SSAP. Cell Mol Biol Lett 12:103–110
Domingues DS, Cruz GM, Metcalfe CJ, Nogueira FT, Vicentini R, Alves CS, Van Sluys MA (2012) Analysis of plant LTR-retrotransposons at the fine-scale family level reveals individual molecular patterns. BMC Genom 13:137. doi:10.1186/1471-2164-13-137
Doyle J (1990) Isolation of Plant DNA from fresh tissue. Focus 12:13–15
Du J, Tian Z, Hans CS, Laten HM, Cannon SB, Jackson SA, Shoemaker RC, Ma J (2010) Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J 63:584–598
Duke N, Kathiresan K, Salmo III SG, Fernando ES, Peras JR, Sukardjo S, Miyagi T (2010) Kandelia candel. The IUCN red list of threatened species: e.T178857A7629021. doi:10.2305/IUCN.UK.2010-2.RLTS.T178857A7629021.en
El Baidouri M, Panaud O (2013) Comparative genomic paleontology across plant kingdom reveals the dynamics of TE-driven genome evolution. Genome Biol Evol 5:954–965
Ellis TH, Poyser SJ, Knox MR, Vershinin AV, Ambrose MJ (1998) Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Mol Genet Genomics 260:9–19
Finatto T, de Oliveira AC, Chaparro C, da Maia LC, Farias DR, Woyann LG, Mistura CC, Soares-Bresolin AP, Llauro C, Panaud O, Picault N (2015) Abiotic stress and genome dynamics: specific genes and transposable elements response to iron excess in rice. Rice (N Y). doi:10.1186/s12284-015-0045-6
Flavell AJ, Dunbar E, Anderson R, Pearce SR, Hartley R, Kumar A (1992) Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res 20:3639–3644
Geng Q, Lian C, Goto S, Tao J, Kimura M, Islam MS, Hogetsu T (2008) Mating system, pollen and propagule dispersal, and spatial genetic structure in a high-density population of the mangrove tree Kandelia candel. Mol Ecol 17:4724–4739
Grandbastien MA (1998) Activation of plant retrotransposons under stress conditions. Trends Plant Sci 3:181–187
Grandbastien MA, Audeon C, Bonnivard E, Casacuberta JM, Chalhoub B, Costa AP, Le QH, Melayah D, Petit M, Poncet C, Tam SM, Van Sluys MA, Mhiri C (2005) Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet Genome Res 110:229–241
Guan C, Li C, Chen W, Mo R, Zhang Q, Du X, Liu J, Luo Z (2015) SSAP analysis reveals candidate genes associated with deastringency in persimmon (Diospyros kaki Thunb.) treated with 40 °C water. Tree Genet Genomes 11:1–13
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
He P, Ma Y, Zhao G, Dai H, Li H, Chang L, Zhang Z (2010) FaRE1: a transcriptionally active Ty1-copia retrotransposon in strawberry. J Plant Res 123:707–714
Ievina B, Syed NH, Flavell AJ, Ievinsh G, Rostoks N (2010) Development of retrotransposon-based SSAP molecular marker system for study of genetic diversity in sea holly (Eryngium maritimum L.). Plant Genet Resour 8:258–266
Jiang N, Gao D, Xiao H, van der Knaap E (2009) Genome organization of the tomato sun locus and characterization of the unusual retrotransposon Rider. Plant J 60:181–193
Jiang N, Visa S, Wu S, Knaap EVD (2012) Rider transposon insertion and phenotypic change in tomato. Top Curr Genet 24:297–312
Jiao Y, Ma RJ, Shen ZJ, Yu ML (2014) Development of Ty1-copia retrotransposon-based SSAP molecular markers for the study of genetic diversity in peach. Biochem Syst Ecol 57:270–277
Kalendar R, Flavell AJ, Ellis TH, Sjakste T, Moisy C, Schulman AH (2011) Analysis of plant diversity with retrotransposon-based molecular markers. Heredity 106:520–530
Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO (2006) Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6:29
Kiær LP, Felber F, Flavell A, Guadagnuolo R, Guiatti D, Hauser TP, Olivieri AM, Scotti I, Syed N, Vischi M (2009) Spontaneous gene flow and population structure in wild and cultivated chicory, Cichorium intybus L. Genet Resour Crop Evol 56:405–419
Kolano B, Bednara E, Weiss-Schneeweiss H (2013) Isolation and characterization of reverse transcriptase fragments of LTR retrotransposons from the genome of Chenopodium quinoa (Amaranthaceae). Plant Cell Rep 32:1575–1588
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Kumar A, Hirochika H (2001) Applications of retrotransposons as genetic tools in plant biology. Trends Plant Sci 6:127–134
Kumar A, Pearce SR, McLean K, Harrison G, Heslop-Harrison JS, Waugh R, Flavell AJ (1997) The Ty1-copia group of retrotransposons in plants: genomic organisation, evolution, and use as molecular markers. Genetica 100:205–217
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Lewontin RC (1972) The apportionment of human diversity. Evol Biol 6:381–398
Lin X, Peng F, Huang J, Zhang T, Shi S, Tang T (2013) Isolation and characterization of RARE-1, a Ty1/copia-like retrotransposon domesticated in the genome of Rhizophora apiculata. Biochem Syst Ecol 50:248–257
Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101:12404–12410
Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross-Ibarra J, Springer NM (2015) Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet 11:e1004915. doi:10.1371/journal.pgen.1004915
McCue AD, Slotkin RK (2012) Transposable element small RNAs as regulators of gene expression. Trends Genet 28:616–623
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Parisod C, Alix K, Just J, Petit M, Sarilar V, Mhiri C, Ainouche M, Chalhoub B, Grandbastien MA (2010) Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol 186:37–45
Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O (2010) Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22:3118–3129
Rico-Cabanas L, Martinez-Izquierdo JA (2007) CIRE1, a novel transcriptionally active Ty1-copia retrotransposon from Citrus sinensis. Mol Genet Genomics 277:365–377
Rocheta M, Cordeiro J, Oliveira M, Miguel C (2007) PpRT1: the first complete gypsy-like retrotransposon isolated in Pinus pinaster. Planta 225:551–562
Sonnhammer EL, Durbin R (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1–GC10
Sun M, Wong KC, Lee JS (1998) Reproductive biology and population genetic structure of Kandelia candel (Rhizophoraceae), a viviparous mangrove species. Am J Bot 85:1631–1637
Syed NH, Flavell AJ (2006) Sequence-specific amplification polymorphisms (SSAPs): a multi-locus approach for analyzing transposon insertions. Nat Protoc 1:2746–2752
Takeuchi T, Sugaya T, Kanazashi A, Yoshimaru H, Katsuta M (2001) Genetic diversity of Kandelia candel and Bruguiera gymnorrhiza in the Southwest Islands, Japan. J For st Res 6:157–162
Tang T, He L, Peng F, Shi S (2011) Habitat differentiation between estuarine and inland Hibiscus tiliaceus L. (Malvaceae) as revealed by retrotransposon-based SSAP marker. Aust J Bot 59:515–522
Tomlinson PB (1986) The botany of mangroves. Cambridge University Press, Cambridge
Ungerer MC, Strakosh SC, Zhen Y (2006) Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr Biol 16:R872–R873
Ungerer MC, Strakosh SC, Stimpson KM (2009) Proliferation of Ty3/gypsy-like retrotransposons in hybrid sunflower taxa inferred from phylogenetic data. BMC Biol 7:40. doi:10.1186/1741-7007-7-40
Voytas DF, Cummings MP, Koniczny A, Ausubel FM, Rodermel SR (1992) copia-like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci USA 89:7124–7128
Wang H, Liu JS (2008) LTR retrotransposon landscape in Medicago truncatula: more rapid removal than in rice. BMC Genom 9:382. doi:10.1186/1471-2164-9-382
Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BB, Powell W (1997) Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol Genet Genomics 253:687–694
Wicker T, Keller B (2007) Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res 17:1072–1081
Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E (2008) A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319:1527–1530
Yang G, Zhou R, Tang T, Shi S (2008) Simple and efficient isolation of high-quality total RNA from Hibiscus tiliaceus, a mangrove associate and its relatives. Prep Biochem Biotechnol 38:257–264
Yeh FC, R-c Yang, Boyle TB, Ye Z, Ma JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Canada, p 10
Acknowledgments
We thank Zixiao Guo for the population sampling. We thank Jianhua Huang for providing experimental assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the National Science Foundation of China (31170308, 91231117 and 31301010), the Science Foundation for Outstanding Young Teachers in Higher Education of Guangdong (Yq2013005), the Fundamental Research Funds for the Central Universities (16lgjc75), and the Chang Hungta Science Foundation of Sun Yat-sen University.
Conflict of interest
All authors declares that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, W., Wang, Y., Shen, X. et al. Isolation, characterization, and marker utility of KCRE1, a transcriptionally active Ty1/copia retrotransposon from Kandelia candel . Mol Genet Genomics 291, 2031–2042 (2016). https://doi.org/10.1007/s00438-016-1237-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-016-1237-5