Abstract
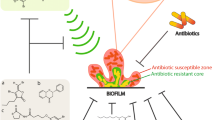
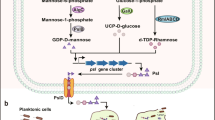
Biofilms are association of microorganisms that attach to each other to a surface enclosed in a self-generated extracellular matrix. Virtually (99.9%) all microorganisms have the competence to form biofilm. The formation of biofilm is a complex process, in which bacterial cells transform from planktonic cells to sessile mode of growth. The biofilm development results from the expression of specific genes. Biofilms have been developed as an adaptive strategy of bacterial species to survive in adverse environmental conditions as well as to establish antagonistic or beneficial interactions with their host. Molecular interaction and details of biofilm formation are not well-understood as bacteria in the biofilm have several orders of magnitude, more resistant to antibiotics compared to planktonic bacteria. Thus, the currently available drugs typically failed to target bacterial biofilms. Quorum sensing (QS) is a process of intercellular signalling or cell-cell communication and a vital regulatory mechanism for coordinating biofilm formation including common activities and physiological processes such as symbiosis, formation of spores or fruiting bodies, antibiotics synthesis, genetic competence, apoptosis and virulence in many bacterial species using extracellular QS signalling molecules, which is often referred to as autoinducers (AIs). Microorganisms produce a wide variety of QS signalling molecules that can be self-recognized in a concentration-dependent manner and subsequently induce or suppress expression of QS-controlled genes. Bacterial QS regulation is established through a wide range of signals such as oligopeptides, N-acyl homoserine lactones (AHLs), furanosyl borate, hydroxy palmitic acid methyl ester and methyldodecanoic acid. In this chapter, we highlight the current understanding of the processes that lead to bacterial biofilm formation, survival behaviours and mechanisms of antimicrobial resistance in bacteria.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Anderson GG, O’Toole GA (2008) Innate and induced resistance mechanisms of bacterial biofilms. In: Romeo T (ed) Bacterial biofilms, Current topics in microbiology and immunology, vol 322. Springer-Verlag, Berlin/Heidelberg, pp 85–105
Arce Miranda JE, Sotomayor CE, Albesa I, Paraje MG (2011) Oxidative and nitrosative stress in Staphylococcus aureus biofilm. FEMS Microbiol Lett 315:23–29
Bagge N, Hentzer M, Andersen JB, Ciofu O, Givskov M, Hoiby N (2004) Dynamics and spatial distribution of beta-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 48:1168–1174
Baronetti JL, Angel Villegas N, Paraje MG, Albesa I (2011) Nitric oxide-mediated apoptosis in rat macrophages subjected to Shiga toxin 2 from Escherichia coli. Microbiol Immunol 55:231–238
Bassler BL (1999) How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol 2:582–587
Bassler BL (2002) Small talk: cell-to-cell communication in bacteria. Cell 109:421–424
Becerra MC, Paez PL, Larovere LE, Albesa I (2006) Lipids and DNA oxidation in Staphylococcus aureus as a con sequence of oxidative stress generated by ciprofloxacin. Mol Cell Biochem 285:29–34
Bjarnsholt T, Jensen PØ, Burmølle M, Hentzer M, Haagensen JAJ, Hougen HP, Calum H, Madsen KG, Moser C, Molin S, Høiby N, Givskov M (2005) Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 151:373–383
Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P (2007) Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator rot by an antisense mechanism. Genes Dev 21:1353–1366
Chadha T (2014) Bacterial biofilms: survival mechanisms and antibiotic resistance. J Bacteriol Parasitol 5:190
Characklis WG, Marshal KC (1990) Biofilms. Wiley, New York
Chen X, Schauder S, Potier N, Van Dorssealaer A, Pelczer I, Bassler BL, Hughson FM (2002) Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322
Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G (2003) The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest 112:1466–1477
Darch SE, West SA, Winzer K, Diggle SP (2012) Density-dependent fitness benefits in quorum sensing bacterial populations. Proc Natl Acad Sci U S A 109:8259–8263
Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG (2004) Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101:1339–1144
Deziel E, Gopalan S, Tampakaki AP, Lépine F, Padfield KE, Saucier M, Xiao G, Rahme LG (2005) The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol 55:998–1014
Dunne WM (2002) Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev 15:155–166
Engebrecht J, Silverman M (1984) Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci U S A 81:4154–4158
Erickson DL, Endersby R, Kirkham A, Stuber K, Vollman DD, Rabin HR, Mitchell I, Storey DG (2002) Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect Immun 70:1783–1790
Federle MJ, Bassler BL (2003) Interspecies communication in bacteria. J Clin Invest 112:1291–1299
Fuqua C, Parsek MR, Greenberg EP (2001) Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35:439–468
Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C (2002) Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol 184:6472–6480
Garrett TR, Bhakoo M, Zhang Z (2008) Bacterial adhesion and biofilms on surfaces. Prog Nat Sci 18:1049–1056
Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP (2006) Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol 61:1038–1048
Hall-Stoodley L, Stoodley P (2002) Developmental regulation of microbial biofilms. Curr Opin Biotechnol 13:228–233
Hancock REW, Siehnel R, Martin N (1990) Outer membrane proteins of Pseudomonas. Mol Microbiol 4:1069–1075
Hausner M, Wuertz S (1999) High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl Environ Microbiol 65:3710–3713
Havarstein LS, Coomaraswamy G, Morrison DA (1995) An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92:11140–11144
Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O (2010) Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332
Irie Y, Parsek MR (2008) Bacterial biofilms. In: Romeo T (ed) Quorum sensing and microbial biofilms. Springer, Berlin/Heidelberg, pp 67–84
Jefferson KK (2004) What drives bacteria to produce a biofilm? FEMS Microbiol Lett 236:163–173
Ji G, Beavis RC, Novick RP (1995) Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci U S A 92:12055–12059
Ji G, Beavis R, Novick RP (1997) Bacterial interference caused by autoinducing peptide variants. Science 276:2027–2030
Jolivet-Gougeon A, Bonnaure-Mallet M (2014) Biofilms as a mechanism of bacterial resistance. Drug Discov Today Technol 11:49–56
Joo HS, Otto M (2012) Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem Biol 19:1503–1513
Keren I, Kaldalu N, Spoering A, Wang YP, Lewis K (2004) Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230:13–18
Le Bars H, Le Gall-David S, Renoux VM, Bonnaure-Mallet M, Jolivet-Gougeon A, Bousarghin L (2012) Impact of a mutator phenotype on motility and cell adherence in Salmonella Heidelberg. Vet Microbiol 159:99–106
Lewis K (2005) Persister cells and the riddle of biofilm survival. Biochemistry 70:267–274
Li YH, Tian X (2012) Quorum sensing and bacterial social interactions in biofilms. Sensors 12:2519–2538
Liu Y, Yang SF, Li Y, Xu H, Qin L, Tay JH (2004) The influence of cell and substratum surface hydrophobicities on microbial attachment. J Biotechnol 110:251–256
Lu TK, Collins JJ (2007) Dispersing biofilms with engineered enzymatic bacteriophage. PNAS 104:11197–11202
Lujan AM, Macia MD, Yang L, Molin S, Oliver A, Smania AM (2011) Evolution and adaptation in Pseudomonas aeruginosa biofilms driven by mismatch repair system-deficient mutators. PLoS One 6:e27842
Lyon GJ, Muir TW (2003) Chemical signaling among bacteria and its inhibition. Chem Biol 10:1007–1021
Martinez JL (2009) The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc Biol Sci 276:2521–2530
Martínez JL (2012) Natural antibiotic resistance and contamination by antibiotic resistance determinants: the two ages in the evolution of resistance to antimicrobials. Front Microbiol 3:1
Mena A, Macia MD, Borrell N, Moya B, de Francisco T, Perez JL, Oliver A (2007) Inactivation of the mismatch repair system in Pseudomonas aeruginosa attenuates virulence but favors persistence of oropharyngeal colonization in cystic fibrosis mice. J Bacteriol 189:3665–3668
Miller MB, Bassler BL (2001) Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199
Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson FM (2004) Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell 15:677–687
Monnet V, Gardan R (2015) Quorum-sensing regulators in Gram-positive bacteria: ‘cherchez le peptide’. Mol Microbiol 97:181–184
Musk DJ Jr, Hergenrother PJ (2006) Chemical countermeasures for the control of bacterial biofilms: effective compounds and promising targets. Curr Med Chem 13:2163–2177
Ng WL, Bassler BL (2009) Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222
Novick RP (2003) Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449
Novick RP, Geisinger E (2008) Quorum sensing in staphylococci. Annu Rev Genet 42:541–564
O’Toole G, Kaplan HB, Kolter R (2000) Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79
Okada M, Sato I, Cho SJ, Iwata H, Nishio T, Dubnau D, Sakagami Y (2005) Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat Chem Biol 1:23–24
Palmer J, Flint S, Brooks J (2007) Bacterial cell attachment, the beginning of a biofilm. J Ind Microbiol Biotechnol 34:577–588
Paraje M (2011) Antimicrobial resistance in biofilms. In: Méndez-Vilas A (ed) Science against microbial pathogens: communicating current research and technological advances. Formatex Research Center, Badajoz, pp 736–744
Parsek MR, Greenberg EP (2005) Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13:27–33
Parsek MR, Singh PK (2003) Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol 57:677–701
Patel R (2005) Biofilms and antimicrobial resistance. Clin Orthop Relat Res 437:41–47
Penesyan A, Gillings M, Paulsen IT (2015) Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules 20:5286–5298
Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH (1999) Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96:11229–11234
Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO (2015) Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med Chem 7:493–512
Rasmussen TB, Givskov M (2006) Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol 296:149–161
Renner LD, Weibel DB (2011) Physicochemical regulation of biofilm formation. MRS Bull 36:347–355
Ruby EG (1996) Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol 50:591–624
Sardesai VM (1995) Role of antioxidants in health maintenance. Nutr Clin Pract 10:19–25
Sauer FG, Remaut H, Hultgren SJ, Waksman G (2004) Fiber assembly by the chaperone-usher pathway. Biochim Biophys Acta 1694:259–267
Schaefer AL, Val DL, Hanzelka BL, Cronan JE Jr, Greenberg EP (1996) Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci U S A 93:9505–9509
Schauder S, Bassler BL (2001) The languages of bacteria. Genes Dev 15:1468–1480
Sekhar S, Kumar R, Chakraborti A (2009) Role of biofilm formation in the persistent colonization of Haemophilus influenzae in children from northern India. J Med Microbiol 58:1428–1432
Sifri CD (2008) Quorum sensing: bacteria talk sense. Healthcare Epidemiol 47:1070–1076
Simon MI, Crane BR, Crane A (2007) Two-component signaling systems. Academic Press, San Diego
Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP (2000) Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764
Solomon JM, Lazazzera BA, Grossman AD (1996) Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev 10:2014–2024
Sordi LD, Muhlschlegal FA (2009) Quorum sensing and fungal-bacterial interactions in Candida albicans: a communication network regulating microbial coexistence and virulence. FEMS Yeast Res 9:990–999
Staley C, Dunny GM, Sadowsky MJ (2014) Environmental and animal associated enterococci. Adv Appl Microbiol 87:147–186
Stevens AM, Dolan KM, Greenberg EP (1994) Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc Natl Acad Sci U S A 91:12619–12623
Sturme MH, Kleerebezem M, Nakayama J, Akkermans AD, Vaugha EE, de Vos WM (2002) Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeuwenhoek 81:233–243
Sutherland IW (1999) Polysaccharases for microbial exopolysaccharides. Carbohydr Polym 38:319–328
Sutherland IW (2001a) The biofilm matrix – an immobilized but dynamic microbial environment. Trends Microbiol 9:222–227
Sutherland I (2001b) Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3–9
Sutherland IW (2001c) Microbial polysaccharides from Gram-negative bacteria. Int Dairy J 11:663–674
Taga ME, Semmelhack JL, Bassler BL (2001) The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol Microbiol 42:777–793
van Bodman SB, Willey JM, Diggle SP (2008) Cell-cell communication in bacteria: united we stand. J Bacteriol 190:4377–4391
Van Delden C, Iglewski BH (1998) Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis 4:551–560
Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR (2005) Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol 3:383–396
Waters CM, Bassler BL (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346
Watnick PI, Kolter R (1999) Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol 34:586–595
West SA, Winzer K, Gardner A, Diggle SP (2012) Quorum sensing and the confusion about diffusion. Trends Microbiol 20:586–594
Williams P, Winzer K, Chan WC, Camara M (2007) Look who’s talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond Ser B Biol Sci 362:1119–1134
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Subramani, R., Jayaprakashvel, M. (2019). Bacterial Quorum Sensing: Biofilm Formation, Survival Behaviour and Antibiotic Resistance. In: Bramhachari, P. (eds) Implication of Quorum Sensing and Biofilm Formation in Medicine, Agriculture and Food Industry . Springer, Singapore. https://doi.org/10.1007/978-981-32-9409-7_3
Download citation
DOI: https://doi.org/10.1007/978-981-32-9409-7_3
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-32-9408-0
Online ISBN: 978-981-32-9409-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)