Abstract
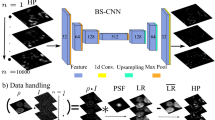
Deconvolution microscopy has been extensively used to improve the resolution of the widefield fluorescent microscopy. Conventional approaches, which usually require the point spread function (PSF) measurement or blind estimation, are however computationally expensive. Recently, CNN based approaches have been explored as a fast and high performance alternative. In this paper, we present a novel unsupervised deep neural network for blind deconvolution based on cycle consistency and PSF modeling layers. In contrast to the recent CNN approaches for similar problem, the explicit PSF modeling layers improve the robustness of the algorithm. Experimental results confirm the efficacy of the algorithm.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Chan, T.F., Wong, C.K.: Total variation blind deconvolution. IEEE Trans. Image Process. 7(3), 370–375 (1998)
Chaudhuri, S., Velmurugan, R., Rameshan, R.: Blind deconvolution methods: a review. In: Blind Image Deconvolution: Methods and Convergence, pp. 37–60. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10485-0_3
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016, Part II. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Goodfellow, I., et al.: Generative adversarial nets. In: Advances in Neural Information Processing Systems, pp. 2672–2680 (2014)
Isola, P., Zhu, J.Y., Zhou, T., Efros, A.A.: Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 1125–1134 (2017)
Kang, E., Koo, H.J., Yang, D.H., Seo, J.B., Ye, J.C.: Cycle-consistent adversarial denoising network for multiphase coronary CT angiography. Med. Phys. 46(2), 550–562 (2019)
Kupyn, O., Budzan, V., Mykhailych, M., Mishkin, D., Matas, J.: DeblurGAN: blind motion deblurring using conditional adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 8183–8192 (2018)
Lu, Y., Tai, Y.W., Tang, C.K.: Conditional CycleGAN for attribute guided face image generation. arXiv preprint: arXiv:1705.09966 (2017)
Mao, X., Shen, C., Yang, Y.B.: Image restoration using very deep convolutional encoder-decoder networks with symmetric skip connections. In: Advances in Neural Information Processing Systems, pp. 2802–2810 (2016)
Mao, X., Li, Q., Xie, H., Lau, R.Y., Wang, Z., Paul Smolley, S.: Least squares generative adversarial networks. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 2794–2802 (2017)
McCann, M.T., Jin, K.H., Unser, M.: Convolutional neural networks for inverse problems in imaging: a review. IEEE Signal Process. Mag. 34(6), 85–95 (2017)
McNally, J.G., Karpova, T., Cooper, J., Conchello, J.A.: Three-dimensional imaging by deconvolution microscopy. Methods 19(3), 373–385 (1999)
Nehme, E., Weiss, L.E., Michaeli, T., Shechtman, Y.: Deep-storm: super-resolution single-molecule microscopy by deep learning. Optica 5(4), 458–464 (2018)
Pizer, S.M., et al.: Adaptive histogram equalization and its variations. Comput. Vis. Graph. Image Process. 39(3), 355–368 (1987)
Rivenson, Y., Göröcs, Z., Günaydin, H., Zhang, Y., Wang, H., Ozcan, A.: Deep learning microscopy. Optica 4(11), 1437–1443 (2017)
Sarder, P., Nehorai, A.: Deconvolution methods for 3-D fluorescence microscopy images. IEEE Signal Process. Mag. 23(3), 32–45 (2006)
Ulyanov, D., Vedaldi, A., Lempitsky, V.: Instance normalization: the missing ingredient for fast stylization. arXiv preprint: arXiv:1607.08022 (2016)
Weigert, M., Royer, L., Jug, F., Myers, G.: Isotropic reconstruction of 3D fluorescence microscopy images using convolutional neural networks. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017, Part II. LNCS, vol. 10434, pp. 126–134. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66185-8_15
You, Y.L., Kaveh, M.: A regularization approach to joint blur identification and image restoration. IEEE Trans. Image Process. 5(3), 416–428 (1996)
Zhu, J.Y., Park, T., Isola, P., Efros, A.A.: Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 2223–2232 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Lim, S., Ye, J.C. (2019). Blind Deconvolution Microscopy Using Cycle Consistent CNN with Explicit PSF Layer. In: Knoll, F., Maier, A., Rueckert, D., Ye, J. (eds) Machine Learning for Medical Image Reconstruction. MLMIR 2019. Lecture Notes in Computer Science(), vol 11905. Springer, Cham. https://doi.org/10.1007/978-3-030-33843-5_16
Download citation
DOI: https://doi.org/10.1007/978-3-030-33843-5_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-33842-8
Online ISBN: 978-3-030-33843-5
eBook Packages: Computer ScienceComputer Science (R0)