Abstract
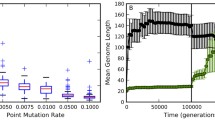
In this paper we investigate the evolutionary pressures influencing genome size in artificial organisms. These were designed with three organisation levels (genome, proteome, phenotype) and are submitted to local mutations as well as rearrangements of the genomic structure. Experiments with various per-locus mutation rates show that the genome size always stabilises, although the fitness computation does not penalise genome length. The equilibrium value is closely dependent on the mutational pressure, resulting in a constant genome-wide mutation rate and a constant average impact of rearrangements. Genome size therefore self-adapts to the variation intensity, reflecting a balance between at least two pressures: evolving more and more complex functions with more and more genes, and preserving genome robustness by keeping it small.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Maynard-Smith, J.: Overview - unsolved evolutionary problems. In: Dover, G.A., Flavell, R.B. (eds.) Genome evolution, pp. 375–382. Academic Press, New York (1982)
Lenski, R.E.: Phenotypic and genomic evolution during a 20,000-generation experiment with the bacterium Escherichia coli. Plant Breeding Reviews 24, 225–265 (2004)
Goldberg, D.E., Deb, K., Kargupta, H., Harik, G.: Rapid accurate optimization of difficult problems using fast messy genetic algorithms. In: Forrest, S. (ed.) Proceedings of the Fifth International Conference on Genetic Algorithms, pp. 56–64. Morgan Kaufmann, San Mateo (1993)
Burke, D.S., De Jong, K.A., Grefenstette, J.J., Ramsey, C.L., Wu, A.S.: Putting more genetics into genetic algorithms. Evolutionary Computation 6, 387–410 (1998)
Ochoa, G., Harvey, I., Buxton, H.: Optimal mutation rates ans selection pressure in genetic algorithms. In: Proceedings of Genetic and Evolutionary Computation Conference (GECCO 2000). Morgan Kaufmann, San Francisco (2000)
Eigen, M., Schuster, P.: The hypercycle: A principle of natural self-organization. Springer, Heidelberg (1979)
Petrov, D.A.: Mutational equilibrium model of genome size evolution. Theoretical Population Biology 61, 533–546 (2002)
Mira, A., Ochman, H., Moran, N.A.: Deletional bias and the evolution of bacterial genomes. Trends in Genetics 17, 589–596 (2001)
Gregory, T.R.: Coincidence, coevolution, or causation? dna content, cell size, and the C-value enigma. Biological Reviews of the Cambridge Philosophical Society 76, 65–101 (2001)
Doolittle, W.F., Sapienza, C.: Selfish genes,the phenotype paradigm and genome evolution. Nature 284, 601–603 (1980)
Ohno, S.: So much junk dna in our genome. In: Smith, H.H. (ed.) Evolution of Genetic Systems, New York, Gordon, Breach, pp. 336–370 (1972)
Eigen, M., McCaskill, J., Schuster, P.: The molecular quasi-species. Adv. Chem. Phys. 75, 149–263 (1989)
Van Nimwegen, E., Crutchfield, J.P., Huynen, M.: Neutral evolution of mutational robustness. Proc. Natl. Acad. Sci. USA 96, 9716–9720 (1999)
Ofria, C., Wilke, C.: Avida: A software platform for research in computational evolutionary biology. Artificial Life 10, 191–229 (2004)
Wilke, C.O., Wang, J.L., Ofria, C., Lenski, R.E., Adami, C.: Evolution of digital organisms at high mutation rates leads to the survival of the flattest. Nature 412, 331–333 (2001)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2005 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Knibbe, C., Beslon, G., Lefort, V., Chaudier, F., Fayard, J.M. (2005). Self-adaptation of Genome Size in Artificial Organisms. In: Capcarrère, M.S., Freitas, A.A., Bentley, P.J., Johnson, C.G., Timmis, J. (eds) Advances in Artificial Life. ECAL 2005. Lecture Notes in Computer Science(), vol 3630. Springer, Berlin, Heidelberg. https://doi.org/10.1007/11553090_43
Download citation
DOI: https://doi.org/10.1007/11553090_43
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-540-28848-0
Online ISBN: 978-3-540-31816-3
eBook Packages: Computer ScienceComputer Science (R0)