Abstract
Background
Genus Vicia is a member of family Fabaceae and comprises 180 to 210 species. The most important species is faba bean (Vicia faba) which is still one of the most favourable grain legumes over all the world. The genus contains some additional food crops and a number of forage plants and some other weedy strains such as Vicia angustifolia and Vicia cordata. The aim of the present investigation is to elucidate the phylogenetic relationships among four Vicia species, two species (Vicia angustifolia L. ssp. Angustifolia (2n = 12) and Vicia cordata wulfen ex Hoppe (2n = 10)) belong to section Vicia, Vicia dalmatica A. Kern (2n = 12, section Cracca), and Vicia johannis tamamsch (2n = 14, section Faba).
Results
Two tools have been applied to identify the genetic relationships among the examined species, double fluorescence in situ hybridization (FISH) has been used to localize the sites of 5S and 45S rDNA, and sodium dodecyl sulfate-poly acrylamide gel electrophoretic (SDS-PAGE) patterns of total seed storage protein fractions. Double FISH experiment has not shown any variation in the loci number, but the positions along the chromosomes were different; both Vicia johannis and Vicia dalmatica exhibited the same interstitial 45S rRNA gene loci, while Vicia angustifolia and Vicia cordata have shown single large stretched 45S rRNA loci almost at the terminal region of the shortest chromosome. It could be concluded from the similarity matrix among the Vicia species as computed according to Jaccard coefficient from the SDS-PAGE, that V. cordata is similar to V. angustifolia and V. dalmatica by a percentage of 73 and 69%, respectively, and the most related species to V. johannis is V. dalmatica (~ 64%).
Conclusion
FISH and SDS-PAGE of the total seed storage proteins together reflected the similar genetic relationship among the studied species as fellows, V. angustifolia is more related to V. cordata then comes V. dalmatica and then V. johannis which is at a distal position from the other species.
Similar content being viewed by others
Background
Vicia is a member of the tribe Vicieae (family Fabaceae) and considered a medium-sized genus; the number of species can be estimated as 180 to 210 [1]. This genus has a considerable economic importance species especially Vicia faba and Vicia ervilia, since faba bean is still one of the most favourable grain legumes in the moderate regions of all the world. In addition, the genus contains some additional minor food crops and more than a dozen forage plants, such as the most important common vetch, V. sativa, other species have developed as weedy strains (Vicia angustifolia and Vicia cordata). The genus is distributed almost over all the world mainly from Asia, Europe, and North America to moderate regions of South America and tropical Africa and mainly located in Mediterranean and Irano-Turanian regions [2, 3]
Vicia species were an excellent objection for karyological, cytogenetical, and molecular-genetic studies, especially the very large chromosomes of V. faba which are suitable for such investigations. The karyological characters of this genus exhibit slight variability, especially in regard to the morphology of chromosomes and the DNA content of the genome. The basic chromosome numbers in genus Vicia are X = 5, 6, and 7, and the great majority of the species are diploid and only a small proportion of 4x and even 6x cytotypes have been observed. However, it has been found in a few cases supernumerary chromosomes (B-chromosomes) in cytotypes of Vicia ohwiana with 2n = 12 + (0-2B) and Vicia alpestris with 2n = 28 + (0-1B), evolutionary implication of such chromosomal deviations is still unknown [4,5,6,7].
Fluorescence in situ hybridization investigations consuming rDNA genes as probes have been used in most of plant species [8,9,10,11]. The ribosomal DNA repeat units occur at one or more rRNA gene loci in the genome, the 18S–5.8S–25S rDNA scattered in tandem arrays in one or more chromosomal regions known as a secondary constriction or nucleolar organizing regions (NORs), whereas 5S rDNA is usually independent scattered in one or several regions [8, 12,13,14]. Therefore, rDNA-FISH has been used as an excellent tool to find out the chromosomal evolution within and between related species by recognizing and understanding the chromosomal organization and analysing the genetic relationships. Application of double or multiple probe FISH enables the discrimination of the homologous chromosomes and explores the structural genome changes among many species. It helps to clarify genetic maps and to assign linkage groups to physically marked chromosomes as well [15,16,17,18]. Florescence in situ hybridization has been applied in different Vicia species [19,20,21,22,23,24,25].
Isozymes systems and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) as molecular marker tool still efficient for germplasm characterization and had been successfully used to evaluate the genetic diversity. Seed storage protein electrophoresis as an additional SDS-PAGE tool has been applied to elucidate and trace back the evolution of numerous groups of plant species and genera [26,27,28,29,30,31,32]. The proteins in the seeds of legumes are ranging from about 20% in pea (Pisum sativum L.) and beans (Phaseolus spp.), increasing up to 40% in soybean (Glycine max (L.) Merr.) and lupin (Lupinus spp.) [33].
Seed storage proteins have been studied in several plant species for providing appropriate biological system for the evolution studies. Their patterns could be used to elucidate the degree of diversity between the different species or to discriminate the individuals within the same species [34,35,36,37,38], due to their stability during the course of evolution and are slightly affected by environmental conditions and seasonal changes [39, 40].
The main goal of the present study was to locate the sites of 5S and 45S rDNA as probes by fluorescence in situ hybridization tool on the metaphase chromosomes of four Vicia species (Vicia angustifolia L. ssp. angustifolia, Vicia cordata wulfen ex Hoppe, Vicia dalmatica A. Kern, and Vicia johannis tamamsch) and by SDS-electrophoretic patterns of total seed storage protein fractions to find out the relationships among them.
Methods
Plant material
Four Vicia species were obtained from the germplasm collection (Genebank for agricultural and horticultural crops) of the Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Germany. Vicia johannis tamamsch (accession no. VIC NAR 46/83) belongs to section Faba with 2n = 14 chromosomes, and two species with 2n = 12, Vicia angustifolia L. ssp. angustifolia (accession no. VIC 1178) belongs to section Vicia and Vicia dalmatica A. Kern (accession no. VIC 38/93) belongs to section Cracca, while the fourth one, Vicia cordata wulfen ex Hoppe (accession no. VIC 453) has 2n = 10 chromosomes and belongs to section Vicia.
Chromosome preparation
Chromosome preparations from root tips and FISH were done according to [41] with minor modifications. Seeds were sown on two layers of moistened filter paper in a petri dish and kept in the dark at 25 °C for 2 days. The young germinated root tips were cut and treated with 0.02% aqueous 8-hydroxyquinoline for 3 h at 15 °C and then washed three times with sterile water before fixation in freshly prepared chloroform-acetic acid-ethanol (6:3:1) then in acetic acid-ethanol (1:3) and stored in ethanol 70%.
Fluorescence in situ hybridization
The A. thaliana BAC clone T15P10 (AF167571) bearing the 45S rDNA sequence was labelled with digoxigenin by nick translation, and the 5S rDNA probe was amplified from genomic DNA of A. thaliana and labelled with biotin by PCR with primers specific for the coding region [42]. The biotinylated 5S rDNA was detected by avidin~Texas Red (Vector Laboratories) and amplified by biotinylated goat anti-avidin (Vector Laboratories) and avidin~Texas Red. Digoxigenin-labelled probes were detected by mouse anti-digoxigenin (Jackson ImmunoResearch Laboratories) and goat anti-mouse antibodies conjugated with Alexa 488 (Molecular Probes). The chromosomes were counterstained with 4′,6-diamidin-2-phenylindol (DAPI, 2 μg/ml). The images were captured with a Zeiss Axioplan 2 epifluorescence microscope equipped with a Spot 2e CCD camera. Images were pseudo-coloured and merged using Adobe Photoshop CS software (Adobe).
The classification and numbering of the mitotic chromosomes (Fig. 1) have been done manually for each species from several images which are taken by the microscope. By magnifying the chromosome images in Adobe Photoshop CS software (Adobe) to enlarge the image to the size in which the difference in chromosome size could be clarified, and then arranging the number of these chromosomes according to their decreasing in the size, taking in account the homologous chromosomes which bear the 45S and 5S rRNA genes.
Mitotic metaphase chromosomes of four Vicia species after FISH with rDNA probes; 18S-25S rDNA probe was detected by FITC (green signals) and 5S rDNA probe by Texas red (red signals). The chromosomes were counterstained with DAPI. aVicia johannis (2n = 14). bVicia angustifolia (2n = 12). cVicia dalmatica 2n = 12. dVicia cordata (2n = 10). Bar = 5.0 μm
Proteins profile using SDS-PAGE
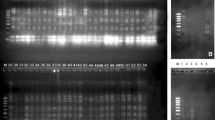
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the method of [43] as modified by [44]. Seed storage proteins (total proteins) were extracted from seeds of different Vicia species. The marker of used protein was BLUltra Prestained Protein Ladder (GeneDirex, cat no. PM001-0500). In this method, 10% protein separating gel were used. Protein fractionations were performed exclusively on vertical slab gel (19.8 cm × 26.8 cm × 0.2 cm) using the electrophoresis apparatus manufactured by Cleaver, UK. The images were captured by a digital camera (Sony, Japan) and transferred directly to the computer; then, the protein bands were analysed by Total Lab programme to find out the molecular weight of each band, subsequently compare the presence and absence of the band among studied species and these data were imported in MVSP (Multi-Variant Statistical Package [45]) to find the similarity matrix and dendrogram (unweighted pair group method with arithmetic (UPGMA), using Jaccardʼs coefficient) which reflect the relationships among the studied species.
Results
rDNA loci
Chromosome number of the analysed species is Vicia johannis (2n = 14), Vicia angustifolia (2n = 12), Vicia dalmatica (2n = 12), and Vicia cordata (2n = 10). Despite double FISH experiment using 45S and 5S rDNA probes has not shown any variation in the number of loci, where one 45S rRNA and one 5S rRNA gene loci were detected in the metaphase chromosomes of each of studied species, but the positions along the chromosomes were different, both Vicia johannis and Vicia dalmatica exhibited the same interstitial 45S rRNA gene loci, while Vicia angustifolia and Vicia cordata have shown single large stretched 45S rRNA loci almost at the terminal region of the shortest chromosome (Fig. 1). These results have been discussed as follows:
Vicia johannis (2n = 14, section Faba) has bigger chromosomes comparing with the other three species, one stretched interstitial 45S rRNA gene site was observed in this species on a smallest chromosome pair (no. 7), which is distinguished with a big satellite, whereas single interstitial 5S rDNA gene site was detected on the short arm of chromosome pair no. 6 (Fig. 1a).
Vicia angustifolia (2n = 12, section Vicia), single large 45S rDNA site, stretched and predominantly located in the terminal region of the smallest chromosome pair (no. 6), while 5S rDNA site was located at subterminal position on the short arm of chromosome pair no. 3 (Fig. 1b).
Vicia dalmatica (2n = 12, section Cracca) has one large stretched interstitial 45S rDNA site was located on the long arm of middle-sized chromosome pair (no. 3), while the 5S rDNA site was at proximal position on the long arm of large chromosome pair no. 2 (Fig. 1c).
Vicia cordata has the lowest chromosome number among the studied species (2n = 10, section Vicia) and is characterized with single terminal 45S rDNA site, and 5S rDNA sites were located separately on the opposite arms of the smallest chromosome pair no. 5 (Fig. 1d).
SDS-electrophoretic patterns
SDS-electrophoretic pattern of total seed storage protein fractions in the studied Vicia species reflected a total number of 44 bands with molecular weight ranging from 210 to 16 Kilo Daltons (KD). The distribution of seed storage protein bands in the four Vicia species is shown in Table 1 and Fig. 2. A maximum number of 35 bands were detected in Vicia cordata, whereas the minimum number was 29 bands in Vicia angustifolia. Although the number of detected bands in Vicia dalmatica and Vicia johannis was 31 and 30 bands, respectively, the monomorphic bands were sixteen at Mw 175, 100, 90, 80, 67, 63, 49, 47, 44, 36, 35, 31, 27, 19, 18, and 17 KD.
Vicia angustifolia exhibited the lowest number of bands (29 bands) among the examined species. The largest size band was at Mw 210 KD and the smallest band was at 17 KD. This species was characterized by having one +ve unique band at Mw 120 KD, and three –ve unique bands at Mw 71, 33, and 32 KD. This species shared with Vicia cordata in 2 bands (130 and 28 KD).
The maximum number of bands (35 bands) was detected in Vicia cordata. The largest-size band was at Mw 210 KD and the smallest band was at 16 KD. This species was characterized by the presence of one +ve unique band at 46 KD, and one –ve unique band at 20 KD. This species shared with Vicia angustifolia in 2 bands (130 and 28 KD), Vicia dalmatica in 3 bands (115, 37, and 23 KD), and Vicia johannis in one band at 16 KD.
Vicia dalmatica had a total number of 31 bands. The largest size band was observed at Mw 175 KD and the smallest band was detected at Mw 17 KD. It was characterized by the presence of one unique band at 62 KD. It shared with Vicia cordata in 3 bands (115, 37, and 23 KD) and Vicia johannis in 2 bands (75 and 41 KD).
In Vicia johannis, a total number of 30 bands was detected, the highest one at 175 KD, whereas the lowest one was at 16 KD. This species was characterized by the presence of four unique bands at Mw 150, 140, 110, and 21 KD.
The similarity indices among these species were estimated for each pair-wise group (Table 2 and Fig. 3). The highest similarity index (0.73) was recorded between Vicia cordata and Vicia angustifolia, while the lowest similarity index (0.51) was recorded between Vicia johannis and Vicia angustifolia. The dendrogram gave two main genetic clusters; the first cluster includes the species Vicia johannis, while the second cluster includes all other studied species. The second cluster was further divided into two sub-clusters; the first sub-cluster includes Vicia dalmatica only, while the second sub-cluster includes Vicia cordata and Vicia angustifolia (Fig. 3).
Discussion
Family Fabaceae is one of the world’s three largest families. It comprises approximately 650 genera. Vicia as one the Fabaceae genera is represented by more than 170 species in the world, and is distributed mainly from North America, Asia, and Europe to the moderate regions of South America and tropical Africa. The genus has economic importance species especially the faba bean (V. faba) as one of the most favourable grain legumes over all the world and V. ervilia as well, in addition to some minor food crops and a number of forage plants, such as V. sativa, and V. angustifolia and V. cordata have developed as weedy strains [2, 3]. Vicia species have a basic chromosome ranged from x = 5, 6 to 7 [4, 6, 7], and only six of them are polyploid [5].
Cytogenetic approaches introduced a powerful system for tracing the organization of DNA sequences along the chromosome. Fluorescence in situ hybridization (FISH) is the most valuable technique in cytogenetic field, it was established more than 40 years ago and has been used to explain all inquiries correlated to mutation, structure changing, and evolution of individual chromosomes and the entire genomes as well. FISH has served as an important tool for chromosome identification in many plant species The 45S rDNA loci consist of tandem arrays of repeating units of the 18S, 5.8S, and 25S rRNA genes, up to thousands of copies of the repeat units may be present in plants, in addition to the 5S rRNA gene repeats (which occurs independent of the 45S rDNA) which are located at one or more locus on chromosome set, and their distinctive sites along chromosomes are providing valuable markers for chromosome identification and offering advantages in comparing different accessions or species [11, 46,47,48].
Molecular cytogenetic approaches like florescence in situ hybridization (FISH) have been applied in different Vicia species [23, 24, 49,50,51]. Faba bean (Vicia faba) is the most extensively studied species via in situ hybridization tool to localize the distribution of repetitive sequences and 5S and 45S rDNA on its chromosomes [25, 52,53,54,55,56,57, 24]. studied FISH with multiple repeated DNA (four BamHI classes) isolated from V. faba and probes of 5S rDNA and 25S rDNA. The authors analysed the chromosomal and genetic relationship between faba bean (Vicia faba) and the closely related wild species (V. johannis). The chromosomal sites of 5S rDNA and 25S rRNA genes could discriminate V. faba from its closely related species. Applied FISH to the chromosomes showed the existence of BamHI family sequences spread in heterochromatin and euchromatin of the V. faba, V. narbonensis, V. hyaeniscyamus, V. galilaea, V. johannis, and V. bithynica.
Florescence in situ hybridization (FISH) has been applied on Vicia species; the study [24] was a trial to find out the physical maps of the 5S rDNA and 25S rDNA sites on the chromosomes of various taxa within genus Vicia (Narbonensis, Villosa, and Sativa), it was the only publication which discussed the positions of 5S rDNA and 45S rDNA by FISH in three species under the current study (Vicia johannis, V. angustifolia, and V. cordata). The current study observations were in agreement with the result obtained by [24] concerning the loci of 45S rDNA and 5S rDNA on the opposite arms of the smallest chromosome pair (no. 5) in Vicia cordata (2n = 10). On the other hand, the current investigation is partially disagreed with the study [24] concerning Vicia johannis (2n = 14), where the locus of 45S rDNA was on the shortest chromosome (no. 7) in both studies, while the locus of 5S rDNA on chromosome pairs no. 3 in their observation, but in the existing study is on chromosome pairs no. 6. Regarding V. angustifolia, the present study is disagreed with them, in their study the loci of 5S rDNA and 45S rDNA were on chromosome pairs no. 4 and 5, respectively, while in the current investigation are on chromosome pairs no. 3 and 6, respectively.
In the present double FISH experiment using 45S and 5S rDNA probes, the result has not shown any variation in the number of loci, where one 45S rDNA and one 5S rDNA gene loci were detected in the early metaphase chromosomes of each of the studied species, but the positions along the chromosomes were different, both Vicia johannis (2n = 14, section Faba) and Vicia dalmatica (2n = 12, section Cracca) exhibited the same interstitial 45S rDNA gene loci, while Vicia angustifolia (2n = 12, section Vicia) and Vicia cordata (2n = 10, section Vicia) have shown single large stretched 45S rDNA loci almost at the terminal region of the shortest chromosome.
Protein fractions in legumes are powerful candidates for improving many food stuffs, such as bakery, meat, and dairy products [58]. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was used to investigate the seed storage protein patterns in many legumes including Vicia in the same study [59], who analysed the seed protein patterns of 47 accessions belonging to 11 species and four tribes of legumes including Vicia. Another study [60] isolated the protein samples from 15 legume species and cultivars (vetches, pea, white lupin, and field bean) and analysed them by SDS-PAGE electrophoresis of the seed storage proteins and estimated their molecular weight and quantify their relative quantities.
Despite this, there were many other evolutionary studies to clarify the relationships among Vicia species by determining the electrophoretic seed proteins patterns [51, 61,62,63,64,65,66,67,68]. It was noticeable that most of previous systematic classification and genetic relationship studies on genus Vicia using SDS-PAGE have been concentrated on faba bean [69,70,71,72,73], which indicates that SDS-PAGE was a useful tool for genetic diversity analysis of faba bean.
There was no previous investigation referred to the species under the present study concerning SDS-PAGE. In the current study, seed proteins of four Vicia species were analysed by SDS-PAGE to evaluate the genetic diversity among them. Eighty different bands of proteins were identified in these species. Depending on the obtained data in Table 2 which shows the similarity matrix among them as computed according to Jaccard coefficient from the SDS-PAGE, it could be concluded that V. cordata is similar to V. angustifolia and V. dalmatica by a percentage of 73 and 69%, respectively, and the most related species to V. johannis is V. dalmatica (~ 64%).
By combining the whole collected results in the present study, depending on double FISH of rDNA gene loci, SDS-PAGE of seed storage protein, in addition to the previously known taxonomical and chromosome number information regarding the species under the current study, the results could be summarized as follows: On one hand, the two species which belong to section Vicia, V. cordata (2n = 10) and V. angustifolia (2n = 12) are the most related to each other by a percentage of 73% and both have shown single large stretched 45S rDNA loci almost at the terminal region of the shortest chromosome. On the other hand V. cordata was similar to V. dalmatica by a percentage of 69%. From another side, V. dalmatica (section Cracca) was similar to V. angustifolia by a percentage of 58%, in the chromosome number (2n = 12) and almost in chromosome size as well, and it was observed that the most related species to V. johannis (section Faba, 2n = 14) is V. dalmatica (56%) and both exhibited the same large interstitial 45S rDNA gene loci.
Finally, according to these explanations, it could be concluded that the genetic relationship among the studied species is that V. angustifolia is more related to V. cordata then comes V. dalmatica and then V. johannis at a literal position from the other species.
Conclusion
Cytogenetic approaches like FISH offer a powerful tool for detecting the organization of highly repeated DNA sequences (e.g., 45S and 5S rRNA genes) on the chromosome by using in situ hybridization of labelled probe sequences directly on the chromosomes. The 45S rRNA with the 5S rRNA genes (occurring as independent tandem arrays of repeating units) are located at one or more sites on the chromosome set, and their distinctive positions on the chromosomes afford useful markers for chromosome identification. The seed storage proteins are characterized by a high degree of polymorphism and limited environmental effect on their electrophoretic patterns; therefore, they have been successfully used as genetic markers for genetic diversity. Traditionally, SDS-PAGE has been used for protein separation and molecular weight determination as a qualitative tool for the analysis of seed legume in the breeding programme (e.g., nutritional qualities of bread and insect resistance) and for the purpose of varietal identification. The current results testified that FISH approach and storage protein profile have successfully reflected the relationships between the studied species.
Availability of data and materials
The authors declare that all generated and analysed data are included in the article. All plant materials (different Vicia species seeds) were identified and collected in the Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Germany.
Abbreviations
- FISH:
-
Fluorescence in situ hybridization
- SDS-PAGE:
-
Sodium dodecyl sulfate-poly acrylamide gel electrophoretic
- NORs:
-
Nucleolar organizing regions
- DAPI:
-
4′,6-Diamidin-2-phenylindol
- MVSP:
-
Multi-Variant Statistical Package
- UPGMA:
-
Unweighted pair group method with arithmetic
- rDNA:
-
Ribosomal DNA
References
Allkin R, Goyder DJ, Bisby FA, White RJ (1986) Names and synonyms of species and subspecies in the Vicieae: issue 3. Vicieae Database Project, Experimental Taxonomic Information Products, Publication 7. Southampton University, Southampton.
Kupicha FK (1981) Vicieae. In: Advances in legume systematics (eds. Polhill, R.M. and Raven, P.M.). Royal Botanic Gardens, Kew, pp 377–381
Maxted N (1993) A phenetic investigation of Vicia (Leguminosae, Vicieae). Bot J Linn Soc 111:155–182
Maxted N, Callimassia A, Bennett MD (1991) Cytotaxonomic studies of Eastern Mediterranean Vicia species (Leguminosae). Pl Syst Evol 177; 221–234 10.1007/B F00937959.
Cremonini R, Funari S, Mazzuca S (1992) Cytology of Vicia species: nuclear structure, karyological analysis and DNA content. Chromatin 1:135–146
Venora G, Ravalli C, Cremonini R (2008) The karyotype as a tool to identify plant species: Vicia species belonging to Vicia subgenus. Caryologia 61:300–319
Arslan E, Ertuğrul K, Öztürk AB (2012) Karyological studies of some species of the genus Vicia L. (Leguminosae) in Turkey, Caryologia, 65:106-113, DOI: 10.1080/000 87114.2012.709804.
Schwarzacher T (2003) DNA, chromosomes, and in situ hybridization. Genome 46:953–962
Garcia S, Panero JL, Siroky J, Kovařík A (2010) Repeated reunions and splits feature the highly dynamic evolution of 5S and 35S ribosomal RNA genes rDNA in the Asteraceae family. BMC Plant Biol 10, 176. doi: 10.1186/1471-2229-10-176
Garcia S, Garnatje T, Kovařík, A (2012) Plant rDNA database: ribosomal DNA loci information goes online. Chromosoma 121: 389–394 10.1007/s00412-012-0368-7.
Jiang J (2019) Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosome Res 27: 153–165. 10.1007/s10577-019-09607-z
Leitch AR, Heslop-Harrison J (1993) Ribosomal RNA gene expression and localization in cereals. In: Chromosomes Today. Springer, Dordrecht.
Lakshmanan PS, Van Laere K, Eeckhaut T, Van Huylenbroeck J, Van Bockstaele E, Khrustaleva L (2015) Karyotype analysis and visualization of 45S rRNA genes using fluorescence in situ hybridization in aroids (Araceae). Comparative Cytogenetics 9:145–160.
Vasconcelos EV, Vasconcelos S, Ribeiro T, Benko-Iseppon AM, Brasileiro-Vidal AC (2018) Karyotype heterogeneity in Philodendron s.l. (Araceae) revealed by chromosome mapping of rDNA loci. PLoS ONE 13(11): e0207318. 10.1371/journal.pone.0207318
Schubert I (2007) Chromosome evolution. Current Opinion in Plant Biology 10: 09–115. doi: 10.1016/j.pbi.2007.01.001
Souza LGR, Crosa O, Guerra M (2010) Karyological circumscription of Ipheion Rafinesque (Gilliesioideae, Alliaceae). Plant Systematics and Evolution 287:119–127.
Braz GT, He L, Zhao H, Zhang T, Semrau K, Rouillard JM, Torres GA, Jiang JM (2018) Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics 208:513–523. 10. 1534/genetics.117.300344
Luo X, Liu J (2019) Fluorescence In Situ Hybridization (FISH) Analysis of the locations of the oligonucleotides 5S rDNA, (AGGGTTT)3, and (TTG)6 in three genera of Oleaceae and their phylogenetic framework. Genes, 10(5), 375. 10.3390/ genes 10050375
Hizume M (1992) Exact location of rRNA genes in Vicia faba chromosomes. Cytologia 57:471–475.
Hizume M (1993) Chromosomal localization of 5S rRNA genes in Vicia faba and Crepis capillaris. Cytologia 58:417–421.
Fuchs J, Strehl S, Brandes A, Schweizer D, Schubert I (1998) Molecular-cytogenetic characterization of the Vicia faba genome - heterochromatin differentiation, replication patterns and sequence localization. Chromosome Res 6: 219-230. 10.1023/A:1009215802737
Raina S, Mukai Y, Kawaguchi K, Goel S, Jain A (2001) Physical mapping of 18S-5.8S-26S and 5S ribosomal RNA gene families in three important vetches (Vicia species) and their allied taxa constituting three species complexes. Theor Appl Genet 103:839–845. https://doi.org/10.1007/s001220100706.
Navrátilová A, Neumann P, Macas J (2003) Karyotype analysis of four Vicia species using in situ hybridization with repetitive sequences. Annals of botany, 91: 921-926. 10.1093/aob/mcg099
Raina SN, Shiran B, Kesavacharyulu K, Mahmoudi A (2010) Fluorescence in situ hybridization with multiple repeated DNA probes applied to the analysis of chromosomal and genomic relationship levels between faba bean (Vicia faba) and its close wild relatives. Caryologia 63:215–222. 10.1080/00087 114.2010. 10589730
Ávila Robledillo, L., Koblížková, A., Novák, P. et al. (2018) Satellite DNA in Vicia faba is characterized by remarkable diversity in its sequence composition, association with centromeres, and replication timing. Sci Rep 8;5838. https://doi.org/10.1038/s41598-018-24196-3.
Ladizinsky, G. & Hymowitz, T. Seed protein electrophoresis in taxonomic and evolutionary studies Theoret Appl. Genetics (1979) 54: 145. 10.1007 /BF00263044
Gilliland TJ, Coll R, CAlsyn E, De Loose M, Van Eijk MJT, Roldán-Ruiz I (2000) Estimating genetic conformity between related ryegrass (Lolium) varieties. 1. Morphology and biochemical characterization. Molecular Breeding 6: 569–580. 10.1023/A:1011361731545
Cooke RJ, Reeves JC (2003) Plant genetic resources and molecular markers: variety registration in a new era. Plant Genetic Resources: 1: 81–87. 10. 1079 /PGR200312
Luo S, Feng J, Pang HM (2004) High-throughput protein analysis by multiplexed sodium dodecyl sulfate capillary gel electrophoresis with UV absorption detection. Journal of Chromatography A 1051: 131-134. 10.1016/j.chroma.2004.06.059
Stoyanova S, Boller B (2010) Seed protein electrophoresis for assessment of genetic variation within genotypes of meadow fescue (Festuca pratensis Huds). Czech J. Genet. Plant Breed 46:75–81.
Singh PK, Shrivastava N, Chaturvedi K, Sharma B, Bhagyawant SS (2016) "Characterization of seed storage proteins from chickpea using 2D electrophoresis coupled with mass spectrometry." Biochemistry Research International, 10.1155/2016 /1049462
Mouzo D, Bernal J, López-Pedrouso M, Franco D, Zapata C (2018) Advances in the biology of seed and vegetative storage proteins based on two-dimensional electrophoresis coupled to mass spectrometry. Molecules 23(10), 2462. 10.3390/ molecules23102462
Ðorpevic V (2009) Perspective of soybean in Europe. Grain Legumes 51:16–17.
Bertozo MR, Valls JFM (2001) Seed storage protein electrophoresis in Arachis pintoi and A. repens (Leguminosae) for evaluating genetic diversity. Genetic Resources and Crop Evolution 48:121–130. https://doi.org/10.1023/A:1011272009831.
Omonhinmin CA, Ogunbodede OO. (2013) Genetic diversity, taxonomy and legumins implications of seed storage protein profiling in Fabaceae. African Journal of Biotechnology 1:36–42. https://doi.org/10.5897/AJB12.2725.
Jiang C, Cheng Z, Zhang C. et al. (2014) Proteomic analysis of seed storage proteins in wild rice species of the Oryza genus. Proteome Sci 12, 51. https://doi.org/10.1186/s12953-014-0051-4.
Alege GO, Abu NE, Sunday CE. (2014) Seed protein electrophoresis of some members of the family Fabaceae. African Journal of Biotechnology 13:3730-3735. doi: 10.5897 /AJB2014.13715
Wan Omar WB, Zakeri HA, Salim JM, Fisall UFM, Amran MA (2019) Genetic diversity of seed storage protein in selected Melastomataceae and Fagaceae from Tasik Kenyir. In: Abdullah M., Mohammad A., Nor Zalipah M., Safiih Lola M. (eds) Greater Kenyir landscapes. Springer, Cham. 10.1007/978-3-319-92264-5_8
Bojórquez-Velázquez E, Barrera-Pacheco A, Espitia-Rangel E et al. (2019) Protein analysis reveals differential accumulation of late embryogenesis abundant and storage proteins in seeds of wild and cultivated amaranth species. BMC Plant Biol 19, 59. 10.1186/s12870-019-1656-7
Gregianini, TS, Winge H (2019) Storage protein variability in natural populations of maté (Ilex paraguariensis) in Brazil. Ciência Rural, 49(2), e20180451. Epub February 18 .10.1590/0103-8478cr20180451
Fransz P, Armstrong S, Alonso-Blanco C, Fischer TC, Torres-Ruiz RA, Jones G (1998) Cytogenetics for the model system Arabidopsis thaliana. Plant J. 13: 867-876. 10.1046/j.1365-313X.1998.00086.x
Gottlob-McHugh SG, Lévesque M, MacKenzie K, Olson M, Yarosh O, Johnson DA (1990) Organization of the 5S rRNA genes in the soybean Glycine max (L.) Merrill and conservation of the 5S rDNA repeat structure in higher plants. Genome 33: 486-494. 10.1139/g90-072
Laemmli UK (1970) Cleavage of structural prteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685. doi:10.1038/227680a0
Studier FW (1973) Analysis of bacteriophage T1 early RNAs and proteins of slab gels. J. Mol Biol 79: 237-248. 10.1016/0022-2836(73)90003-X
Kovach WL (1998) MVSP_ Multi-variant statistical package for windows, ver. 3.0 Kovach computing services: Pentraeth, Wales
Heslop-Harrison JS (2000) Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. Plant Cell 12:617–636. https://doi.org/10.1105/tpc.12.5.617
Volkov RA, Komarova NY, Hemleben V (2007) Ribosomal DNA in plant hybrids: inheritance, rearrangement, expression. Syst Biodivers 5:261–276. 10.1 017/S1477200007002447
Garcia S, Kovarik A, Leitch A, Garnatje T (2017) Cytogentic features of rRNA genes across land plants: analysis of plant rDNA database. Plant J 89:1020–1030. 10.1111/tpj.13442
Fuchs, J., Pich, U., Meister, A. et al. (1994) Differentiation of field bean heterochromatin by in situ hybridization with a repeated FokI sequence. Chromosome Res 2:25–28. https://doi.org/10.1007/BF01539450.
Ali HB, El-Khodary SE, El-Ashry ZM, Osman SA (2009) rDNA loci and DAPI bands reflect the phylogenetic distance between Vicia species. Cytologia 74(4):467–472.
Han Y, Liu Y, Wang H, Liu X (2017) The evolution of Vicia ramuliflora (Fabaceae) at tetraploid and diploid levels revealed with FISH and RAPD. PLoS ONE 12(1): e0170695. doi:10.1371/journal.pone.0170695
Schubert I, Baeumlein H, Wobus U (1978) In situ hybridization of iodinated 5S and 18/25S RNA to Vicia faba metaphase chromosomes. Biol Zentralbl 97:129–135.
Cionini PG, Bassi P, Cremonini R, Cavallini A (1985) Cytological localization of fast renaturing and satellite DNA sequences in Vicia faba. Protoplasma, 124: 106-111. 10.1007/BF01279729
Maggini F, Cremonini R, Zolfino C, Tucci GF, D’ovidio R, Delre V, et al. (1991) Structure and chromosomal localization of DNA sequences related to ribosomal subrepeats in Vicia faba. Chromosoma, 100: 229-234. 10.1007/BF00344156
Rawlins DJ, Highett MI, Shaw PJ (1991). Localization of telomeres in plant interphase nuclei by in situ hybridization and 3D confocal microscopy. Chromosoma 100:424-431. 10.1007/BF00364552
Fuchs J, Joos S, Licheter P, Schubert I (1994) Localization of vicilin genes on field bean chromosome II by fluorescent in situ hybridization. Journal of Heredity 85:487-488. 10.1093/oxfordjournals.jhered.a111506
Fuchs J, Schubert I (1995) Localization of seed protein genes on metaphase chromosomes of Vicia faba via fluorescencein situ hybridization. Chromosome Res 3: 94-100. https:// doi.org/10.1007/BF00710669
Rojas-Hijazo B, Garces MM, Caballero ML, Alloza P, Moneo I (2006) Unsuspected lupin allergens hidden in food. International Archives of Allergy and Immunology 141: 47-50. doi: 10.1159/000094181
Valizadeh M (2001) Seed storage protein profile of grain legumes grown in Iran, using SDS-PAGE. Journal of Agricultural Science and Technology (JAST-IRAN) 3:287–292
Nikolic Z, Djordjevic V, Torbica A, Mikić A (2012) Legumes seed storage proteins characterization by SDS-PAGE and Lab-on-a-Chip electrophoresis. Journal of Food Composition and Analysis 28:75-80. 10.1016/j.jfca.2012.08.005
Ladizinsky G, Waines G (1982) Seed protein polymorphism in Vicia sativa aggregate (Fabaceae). Plant Syst Evol 41:1–4.
Haider AS, El-Shanshoury AR (2000) Variability of storage proteins and esterase isozymes in Vicia sativa subspecies. Biol Plant 43:205–209.
Potokina E, Endo Y, Eggi E, Ohashi H (2003) Electrophoretic patterns of seed proteins in the East Asian Vicia species (Leguminosae) and their systematic utility. J. jap Bot 78:29–37.
Mirali N, El-Khouri S, Rizq F (2007) Genetic diversity and relationships in some Vicia species as determined by SDS-PAGE of seed proteins. Biol Plant 51: 660-666. 10.1007/s10535-007-0139-0
De la Rosa L, Gonzalez JM (2010) The genetic diversity associated with seed proteins in a collection of Spanish underground vetches (Vicia sativa L. subsp. amphicarpa (Dorthes) Asch. et Graebn.). Genetic Resources and Crop Evolution 57:565-573. doi: 10.1007/s10722-009-9494-3
Abdel Khalik K, Al-Gohary Iman (2013) Taxonomic relationships in some Vicia species from Egypt, based on seed morphology and SDS-PAGE of seed proteins. Acta Scientiarum Biological Sciences. 35. 603-611. 10.4025/actascibiolsci.v35i4.19345.
Bechkri S, Medoukali I, Khelifi D (2017) Ecogeographic variability and genetic diversity associated with seed albumins, globulins and prolamins patterns in Vicia taxa from Algeria. Bot Stud 58,27. 10.1186/s40529-017-0177-7
Mahmoud AF, Abd El-Fatah BES. (2020) Genetic diversity studies and identification of molecular and biochemical markers associated with fusarium wilt resistance in cultivated faba bean (Vicia faba). Plant Pathol J. 36(1):11–28. https://doi.org/10.5423/PPJ.OA.04.2019.0119.
Tucci M, Capparelli R, Costa A et al. (1991) Molecular heterogeneity and genetics of Vicia faba seed storage proteins. Theoret Appl Genetics 81: 50-58. 10.1007/ BF00226111
Hou WW, Zhang XJ, Shi JB, Liu YJ (2015) Genetic diversity analysis of faba bean (Vicia faba L.) germplasms using sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Genetics and molecular research: GMR. 14:13945-13953. DOI: 10.4238/2015.october.29.15.
Pradhan A, Sinha SK (2012) Genetic variation in storage proteins and isozymes among Vicia faba L. cultivars of Bihar. VI International conference on legume genetics and genomics, Hyderabad.
Pradhan A, Shahi VK. Sinha SK (2014) Evaluation of genetic diversity in faba bean (Vicia faba L.) genotypes using seed protein and isozymes electrophoresis. Natl Acad Sci Lett 37: 303-309. 10.1007/s40009-014-0244-5
Liu Y, Wu X, Hou W, Li P, Sha W, Tian Y (2017) Structure and function of seed storage proteins in faba bean (Vicia faba L.). 3 Biotech 7(1), 74. Doi:10.1007/s13205-017-0691-z
Acknowledgements
The authors would like to thank the Genetics and Cytology department, National Research Centre, Dokki, Giza, Egypt, for performing all experiment in its laboratory. The authors are grateful to all the researchers whom we cited in this review for their significant and valuable research.
Funding
The research experiment was partially sponsored by Department of Genetics and Cytology, National Research Centre, Cairo, Egypt
Author information
Authors and Affiliations
Contributions
HA performed the FISH experiment part and SO performed the seed storage protein electrophoresis part. HA and SO wrote the manuscript and participated in the data discussion, data analyses, and drafting of the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ali, H.B.M., Osman, S.A. Genetic relationship study of some Vicia species by FISH and total seed storage protein patterns. J Genet Eng Biotechnol 18, 37 (2020). https://doi.org/10.1186/s43141-020-00054-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43141-020-00054-6