Abstract
Background
While a relationship between embryo morphology, developmental ability, and genetic integrity exists, the selection of embryos with higher implantation potential remains a major challenge in assisted reproductive technology (ART). This study investigated blastocyst developmental competence and euploidy status in human embryos that had been classed as too poor quality to transfer (ET) or cryopreserve at the cleavage stage.
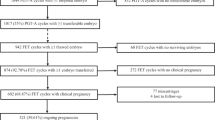
Embryos were divided into three groups. Group 1 (n = 41) included good quality embryos from candidates of preimplantation genetic testing for aneuploidy (PGT-A). Groups II and III were the “rejected” supernumerary embryos, defined as suboptimal for ET or vitrification after morphological examination, with embryos randomly divided between the groups. Group II embryos (n = 31) were cultured up to the day 3 cleavage stage, when they were biopsied and fixed. Group III embryos (n = 27) were cultured up to the day 5 blastocyst stage, when they were evaluated for morphology and chromosomal status. Chromosomal status in all groups was assessed by multi-color fluorescence in situ hybridization (FISH) for chromosomes 13, 18, 21, X, and Y.
Results
Euploidy rates in groups I, II, and III were 56.1%, 38.7%, and 55.5 %, respectively. Among the blastocysts that developed from “rejected” embryos, 59.3% were classed as good quality. The most frequent chromosomal aneuploidy was related to the sex chromosome (22.2%). The mosaicism rate was not significantly different between the group II and III embryos (25.8% vs. 37.0%, p = 0.28).
Conclusion
In conclusion, surplus poor-quality embryos rejected from clinical utilization at the cleavage stage may develop into viable blastocysts with normal chromosomal status for at least 5 chromosomes. Recovery of euploidy during poor-quality embryo transition from cleavage stage to blastocyst could provide an alternative choice for ET.
Similar content being viewed by others
Background
While a relationship between embryo morphology, developmental ability, and genetic integrity exists [1, 2], the selection of embryos with higher implantation potential remains a major challenge in assisted reproductive technology (ART) [3]. Recent studies have shown that the correlation between embryo morphology and implantation potential is relatively weak [1, 4], due to the low predictive value of morphological criteria for embryo selection as a conventional and routine technology in ART clinics [5]. Many IVF laboratories perform embryo transfer (ET) on days 2 or 3 after fertilization. The critical decision of selecting the best embryo for either ET or cryopreservation is commonly taken on the ET day, based on morphological criteria including cell numbers, fragmentation degree, cells size, and cytoplasmic appearance by stereo microscope. Therefore, large proportions of human in vitro generated cleavage-stage embryos may be discarded, due to their poor prognosis for implantation [6]. Nowadays, with improved culture systems, blastocyst production has become more practical and efficient. Studies have also demonstrated that poor-quality cleavage-stage embryos may be capable of implantation [7, 8] and becoming healthy babies [7, 9, 10] when extending the culture to the blastocyst stage. A recent study reported that low-grade day-3 embryos can result in successful blastulation and clinical pregnancies, even if the blastulation rate is slower than normal [8].
In general, human fecundity is considered to be poor compared to other species [11, 12], probably due to the high occurrence of chromosomal abnormalities in early human embryos [13]. The baseline of chromosome abnormalities in human embryos produced through ART was reviewed to be higher than 50%, regardless of maternal age [14]. As well, inverse relationship between advanced maternal age (> 35 years) and embryo euploidy has been known. So that, a comprehensive review on trophectoderm biopsies showed that the rate of embryo aneuploidy increases from age 31 to age 43 at approximately 85% [15].
In addition, it has been postulated that 60% of all IVF day-3 embryos showed at least one aneuploid cell [16].
Preimplantation genetic testing for aneuploidy (PGT-A) and culture of embryos to day 5 may improve ART outcomes by accurately identifying euploid embryos [17]. The importance of this issue becomes more clear when only poor-quality embryos are generated from an ongoing IVF cycle, which might otherwise not be considered for ET or cryopreservation [8]. Therefore, the purpose of the current study was to investigate the morphological as well as cytogenetic assessments of blastocysts, cultured from such surplus poor-quality embryos.
Methods
Patients
This prospective trial was approved by the ethics committee of the Institute for Reproductive Sciences, Yazd, Iran (reference number 175014). Written informed consents were obtained from all patients. Embryos created between August 2015 and February 2017 were prospectively included in the study. The exclusions were patients with a history of recurrent pregnancy loss and genetic disorders, maternal age ≥ 37 years, and severe male factors with non-ejaculated spermatozoa and cycles using donor gametes/embryos.
Ovarian hyperstimulation protocol and oocyte preparation
The ovarian hyperstimulation was performed using the standard GnRH antagonist protocols as described previously [18]. Briefly, 150 IU/day of follicle-stimulating hormone (FSH Gonal F, Serono, Geneva, Switzerland) was administered on day 2 of the menstrual cycle. When at least one follicle reached 13 mm in diameter, 0.25 mg of a GnRH antagonist (Cetrotide, Merck Serono, Darmstadt, Germany) was initiated and continued until the day of human chorionic gonadotropin (hCG) injection. When proper follicular development was viewed on transvaginal ultrasound, recombinant hCG (Ovitrelle, Merck Serono, Germany) was administered to trigger final maturation and ovulation. Approximately 36 h later, ultrasound-guided oocyte collection was performed using a single lumen aspiration needle (Wallace, Cooper Surgical, UK). Retrieved cumulus–oocyte complexes (COCs) were incubated in culture medium (G-IVF; Vitrolife, Kungsbacka, Sweden) covered with mineral oil (Ovoil; Vitrolife) at 37 °C and 6% CO2 for 2–3 h. The COCs were denuded of their cumulus and coronal cells by 30–60 s exposure to HEPES-buffered medium containing 80 IU/ml hyaluronidase (Irvine Scientific, CA, USA) and by pipetting the COCs [19].
Fertilization and embryo assessments
Following ICSI, the oocytes were placed in standard culture dishes containing 1 ml G-1 Medium (G-1; Vitrolife) covered with oil and incubated overnight in a standard incubator (Thermo Scientific, Thermo Fisher Scientific, USA) at 37 °C and 6% CO2. Regularly, fertilized oocytes (2PN) were cultured and embryo morphology was evaluated based on the number of blastomeres and the percentages of fragmentation on days 2 or 3 [20]. Embryos were graded into four categories: grade A, equal size blastomeres and less than 10% fragmentation; grade B, slightly unequal blastomeres with up to 20% fragmentation; grade C, unequal sized blastomeres and up to 50% fragmentation and large granules; and grade D, unequal blastomeres with significant fragmentation (> 50%) and large granules [21]. The best embryos with good quality were transferred, vitrified, or biopsied for PGT-A according to critical treatment cycle defined for each patient. Surplus embryos, considered unsuitable for above processes, were left in the culture for 3 additional days. To increase reproducibility and reliability, two expert embryologists morphologically scored all included embryos based on Hill grading system independently and blindly.
Categorization of embryos
Embryos were divided into three groups depending on their quality and developmental stage (Fig. 1). Good-quality cleavage-stage embryos (grades A and B) were assigned as group I. Also, the good quality embryos in group I, with at least six cells on day 3 were included from the women who were candidates of PGT-A, whereby embryos were transferred into 20 μl of Ca-Mg-free culture medium (Vitrolife, Sweden) under mineral oil, subjected to blastomere biopsy (one nucleated cell). Discarded embryos, defined as suboptimal for ET or cryopreserve, were randomly divided into two groups of II and III using computer-generated random numbers. Group II embryos were cultured for 3 days and at least three of their nucleated blastomeres were biopsied and fixed individually according to previous protocols [22]. Group III embryos were cultured to the blastocyst stage for 5 or 6 days.
Assessment of blastocyst-stage embryos
Group III blastocysts were categorized into the following: grade A, blastocysts were expanded with a distinct inner cell mass (ICM) and many cells organized in the trophectoderm. Grade B, blastocysts were either poorly expanded and/or with less defined ICM and trophectoderm cells, but without signs of degenerative foci. Grade C, the blastocysts exhibited poor morphology, characterized by a number of degenerative foci in the ICM and trophectoderm with a poorly developed blastocyst cavity [23] (Fig. 2). Any group III blastocysts were graded morphologically and underwent trophectoderm biopsy of at least 10–15 cells via blind biopsies for FISH analysis.
Micrograph showing the three different morphology grades (A, B, C) (Hardarson 2003) of blastocysts. Grade A, grade A blastocysts were expanded with a distinct inner cell mass (ICM) and many cells organized in the trophectoderm. Grade B, grade B blastocysts were either poorly expanded and/or with less defined ICM and trophectoderm cells but without signs of degenerative foci. Grade C, blastocysts exhibited poor morphology, characterized by a number of degenerative foci in the ICM and trophectoderm and a poorly developed blastocyst cavity
Fixation and FISH analysis
For the biopsy procedure, the zona was breached using a 1480-nm wavelength infrared diode laser. After biopsy, fixation of nucleated cells was performed [12]. FISH was performed in two rounds. During two rounds of FISH, X and Y chromosome probes, colored by green and red respectively, were used in the first round. Specific probes for chromosomes 13, 18, and 21 detected in green, aqua, and red colors respectively, were used for the second round. The FISH procedure was carried out according to the instructions with slight modifications (Cytocell, OGT Company, Cambridge, UK). Information about enumeration probes were included as [Xp11.1-q11.1 (DXZ1), Yp11.1-q11.1 (DYZ3) and 13q14.2, 21q22.13, 18p11.1-q11.1 (D18Z1)].
For performing the second round, the coverslip was removed at first. The slides were then washed in PBS and dehydrated in increasing ethanol series and hybridized with the second sets of probes. Following hybridization, the slides were counterstained in 4′,6-diamidino-2-phenylindole anti-fade solution. The results of the first and second round FISH were evaluated by two analysts (geneticist and technician), who arrived at a consensus on the diagnosis using a fluorescence microscopy (Olympus BX51 and Genetics GSL-10 with Olympus BX61; Tokyo, Japan). The microscope was equipped with the following filters: triple-band filters (aqua, orange, green) and single-band pass filters (red, green, aqua). Images were captured at × 60 or × 100 magnification using spectral imaging software (Figs. 3 and 4).
Nucleus hybridization of probes with target chromosomes of one blastomere biopsied from cleavage-stage embryo for PGT-A applying FISH technique. The image showed a female normal nucleus for chromosome X (green), chromosome Y (red), and chromosome 18 (blue) in first round as well as chromosome 13 (green) and chromosome 21 (red) in the second round using specific probes (Cytocell, OGT Company, Cambridge, UK) in each round
Preimplantation genetic testing for aneuploidy (PGT-A) of biopsied blastocysts obtained from surplus embryos applying FISH technique. The images showed normal nuclei: a stained with DAPI; b for chromosome X (green), chromosome Y (red), and chromosome 18 (blue) in first round; and c a blastocyst with mosaicism for chromosome 13 (green), and chromosome 18 (blue) using specific probes (Cytocell, OGT Company, Cambridge, UK)
Statistical analysis
Descriptive statistics for continuous data were based on means ± standard deviations or percentages, according to the data normality. Data analysis was performed using SPSS (version 20). The frequency distributions between the groups were analyzed using Fisher’s exact test and Chi-square analysis. Differences were considered significant at p ˂ 0.05.
Results
Embryos at the cleavage (n = 72) and blastocyst (n = 27) stage were analyzed from women (n = 99) aged 28.75 ± 4.11 (group I), 29.35 ± 3.64 (group II), and 30.44 ± 3.30 years (group III). The etiology of infertility was as follows: male factor, 21 (51.2%) vs. 9 (29%) vs. 14 (51.19%); female factor, 10 (24.4%) vs. 12 (38.7%) vs. 8 (52.9%); combined male and female factors, 9 (22%) vs. 7 (22.6%) vs. 0 (0.0%); and for unknown, 1 (2.4%) vs. 3 (9.7%) vs. 5 (18.5%) in groups I, II, and III, respectively.
Table 1 shows the chromosome abnormalities after FISH in the three groups. Of the 41 and 31 embryos at the cleavage-stage in groups I and II, the 56.1% (23/41) and the 38.7% (12/31), respectively, were described as chromosomally euploid for the five chromosomes analyzed, while the 43.9% (18/41) and the 61.3% (19/31) were aneuploid in groups I and II, respectively. Although the aneuploidy rates in discarded embryos (group II) were higher compared to good quality embryos (group I), the differences were insignificant (p ≥ 0.05). The most frequent chromosomal aneuploidy found was the trisomy or monosomy of chromosome 13 (34.1%) in group I as well as the trisomy or monosomy of chromosomes 13 and 18 (29%) in group II and trisomy or monosomy of chromosomes 13 (18.5%) in group III. The highest numbers of sex, autosomal, and total chromosomal abnormalities were detected in group I (24.4%, 10/41), group II (54.8%, 17/31), and group II (61.3%, 19/31), respectively.
The FISH results from 27 blastocysts developed from discarded embryos (group III) are presented in Table 2. The number of examined nuclei ranged from 10 to 15 per embryo. Of 27 discarded embryos that developed to the blastocyst stage, 55.5% (15/27) were euploid, 37.0% (10/27) were mosaic, and 7.4% (2/27) of them had at least one chromosomal abnormality. More than half of the blastocysts that developed from discarded embryos showed good quality in morphological appearance (grade A = 25.9% + grade B = 33.4%), while the poor-quality blastocysts with grade C/D were counted as 40.7%. The morphological grading of blastocysts from group III showed no significant difference as well (p = 0.08). The most frequent aneuploidy belonged to the sex chromosomal abnormalities (22.2%). The mosaicism rates were insignificant between the groups of II and III (25.8% vs. 37 %, respectively; p = 0.28).
Nineteen of 41 (46.3%) embryos in group I, 19 (61.3%) in group II, and 9 (52.9%) in group III were detected as male. There was no difference in the proportion of male and female embryos between the three groups (p = 0.10). In contrast, there was the significant difference in the proportion of male and female embryos in group III (33.3% male vs. 66.7% female, p ˂ 0.001). Also, the distribution of chromosomal aneuploidies regarding sex embryos’ type was significantly different in group III (22.2% male vs. 55.5% female; p ˂ 0.0001).
Discussion
This study compared the aneuploidy rates between good- and poor-quality embryos to assess if the latter can be an additional source of viable embryos for ART patients. No significant difference was observed in aneuploidy rates between embryos with different quality and stages when analyzed by FISH technique. Interestingly, it was also possible to diagnose the presence of mosaicism, if a representative number of blastomere cells were analyzed by cytogenetic analysis [12, 24, 25].
In line with our study, Fesahat and colleagues [26], who investigated the frequency of aneuploidy in five chromosomes from high-quality embryos on day 3, observed that the morphological characterization could not be a reliable method for the embryo selection regarding the considerable chromosomal abnormalities detected in embryos cultured in vitro.
Another study assessed the effect on aneuploidy rates on the morphological appearance of embryos at the cleavage and blastocyst stage [1]. Their results confirmed that chromosomal abnormalities had no visible effect on morphological scores assigned using traditional criteria. They classified the cleavage-stage embryos into three groups (good, average, and poor quality) and reported the aneuploidy rates of 82.4%, 87%, and 90.5%, respectively. However, they showed that some aneuploidies affected the blastocyst morphology during the microscopic observations. It should be noted that they applied comparative genomic hybridization (CGH) microarray for the cytogenetic assessments of human embryos. Similarly, other researchers have analyzed human blastocysts using CGH array and found an aneuploidy rate of 54.4% [27]. Despite some methodological diversity that cannot affect data interpretation, the present findings on aneuploidy rates using FISH were in agreement with other studies.
The novelty of this study was the use of embryos that are usually not selected for ET, due to a poor quality and morphological aspects, as per the embryologist opinion. A total of 27 surplus blastocysts (group III) were analyzed in this study by FISH, with 55.6% having a normal chromosomal status for the five chromosomes assessed and 59.3% having a good morphological appearance. The five chromosomes were selected since not all chromosomes display an equal frequency of aneuploidy and embryos with abnormalities are capable of reaching term [26].
Some studies have shown that blastocysts that are derived from poor-quality cleavage-stage embryos may lead to pregnancy [28, 29]. Kaartinen and colleagues suggested that the use of surplus blastocysts developed from discarded embryos could increase cumulative the delivery rate [10]. However, none of these studies examined the blastocyst euploidy status prior to ET. Genetic assessment is meaningful, when evidence indicates that chromosomally abnormal human embryos are capable of developing to blastocyst stage regardless of their euploidy situation, such as trisomies [30].
One study [2], with the aim of investigating the correlation between blastocyst morphology, euploidy, and implantation, showed a direct association between blastocyst morphology and aneuploidy. In the present study, we could not investigate this relationship because of the limited number of blastocysts. Our data showed that the rate of mosaicism between groups II and III was insignificant (25.8% vs. 37%, respectively). Chromosomal mosaicism is commonly seen in human preimplantation embryos [31] ranging from 4 up to 90% [32].
The FISH technique has been used as the first molecular method for evaluating the chromosomal abnormality. However, a shortcoming of this technique is the lack of assessing whole chromosomes. Therefore, advanced methods like SNP arrays, aCGH, and NGS, have been introduced to overcome FISH limitations. Unfortunately, these techniques also have limitations due to their high costs and subsequent unavailability in many IVF centers [33].
Other studies of human embryos at the early-cleavage stage have reported a mosaicism frequency of 75%, which is consistent with our findings [34]. Others [35, 36] have also reported a high prevalence of mosaicism at the blastocyst stage. This high incidence of mosaicism supports the hypothesis that all human embryos may contain some abnormal cells, but that this may not necessarily impair the development and delivery of a normal child.
Several studies have focused on the implantation and healthy live births after the transfer of mosaic blastocysts [37,38,39,40,41], indicating that that mosaic blastocysts may be self-correcting [42]. Mosaicism in established pregnancies may be derived from the TE cells, rather than the ICM, indicating that mosaic blastocysts could be considered for ET when euploid embryos are not available [33], provided that patients are fully counseled and accepting of the risks.
In line with our results, Huang and colleagues applied CGH and single nucleotide polymorphism microarray (SNPs) for analysis of the small number of embryonic cells at different embryonic developmental stages [43]. They concluded that most embryos maintained chromosomal stability during preimplantation development, with blastocyst-stage embryos providing more reliable aneuploidy results.
While our study did not compare aneuploidy rates in sibling embryos, patients were matched according to demographic status in order to eliminate the selection bias between the groups. Similar to our study, others have compared euploidy between day 3 and day 5 biopsied embryos and concluded that a larger proportion of embryos were euploid on day 5 [44]. Su and colleagues also suggested that aneuploidy rates in day 7 human blastocysts produced by IVF were manifold [9]. However, good quality/euploid blastocysts were eligible for ET on day 7, leading to healthy live births.
Recently, it was shown that the low-quality day-3 embryos can result in successful blastulation and clinical pregnancies. However, the normal blastulation rate was poor [8].
Our study has some limitations, though, including low sample size and use of FISH as an outdated technology. Because of limitation in the budget and equipment in our center, we used this technique for five chromosomes. So we decided on the first step to do FISH for our discarded embryos. Since, we made interesting results, we decided to evaluate the morphokinetic parameters of discarded embryos development via time-lapse monitoring and do more advanced techniques such as array CGH in higher sample size in cooperation with the other center in the next part of our study.
Conclusion
In conclusion, some of surplus poor-quality embryos rejected from clinical utilization at the cleavage stage may develop into viable blastocysts with normal chromosomal status for at least 5 chromosomes. Recovery of euploidy during poor-quality embryo transition from cleavage stage to blastocyst could provide an alternative choice for ET.
Availability of data and materials
The datasets used and/or analyzed during the current study are available.
Abbreviations
- ET:
-
Embryo transfer
- PGT-A:
-
Preimplantation genetic testing for aneuploidy
- ART:
-
Assisted reproductive technology
- HCG:
-
Human chorionic gonadotropin
- ICSI:
-
Intracytoplasmic sperm injection
- IVF:
-
In vitro fertilization
- hCG:
-
Human chorionic gonadotropin
- ICM:
-
Inner cell mass
- TE:
-
Trophectoderm
- COCs:
-
Cumulus–oocyte complexes (COCs)
References
Fragouli E, Alfarawati S, Spath K, Wells D (2013) Morphological and cytogenetic assessment of cleavage and blastocyst stage embryos. Molecular human reproduction. 20(2):117–126
Capalbo A, Rienzi L, Cimadomo D, Maggiulli R, Elliott T, Wright G et al (2014) Correlation between standard blastocyst morphology, euploidy and implantation: an observational study in two centers involving 956 screened blastocysts. Human Reproduction. 29(6):1173–1181
Carrasco B, Arroyo G, Gil Y, Gómez MJ, Rodríguez I, Barri PN et al (2017) Selecting embryos with the highest implantation potential using data mining and decision tree based on classical embryo morphology and morphokinetics. Journal of Assisted Reproduction and Genetics.:1–8
Nogales MDC, Bronet F, Basile N, Martínez EM, Liñán A, Rodrigo L et al (2017) Type of chromosome abnormality affects embryo morphology dynamics. Fertility and sterility 107(1):229–35. e2
Feichtinger M, Vaccari E, Carli L, Wallner E, Mädel U, Figl K et al (2017) Non-invasive preimplantation genetic screening using array comparative genomic hybridization on spent culture media: a proof-of-concept pilot study. Reproductive biomedicine online. 34(6):583–589
Yao G, Xu J, Xin Z, Niu W, Shi S, Jin H et al (2016) Developmental potential of clinically discarded human embryos and associated chromosomal analysis. Scientific reports. 6:23995
Sallem A, Santulli P, Barraud-Lange V, Le Foll N, Ferreux L, Maignien C et al (2018) Extended culture of poor-quality supernumerary embryos improves ART outcomes. Journal of assisted reproduction and genetics. 35(2):311–319
Klement AH, Ovadia M, Wiser A, Berkovitz A, Shavit T, Nemerovsky L et al (2017) What we learned from extended culture of ‘rejected’day-3 cleavage stage embryos: a prospective cohort study. Journal of ovarian research. 10(1):35
Su Y, Li J-J, Wang C, Haddad G, Wang W-H (2016) Aneuploidy analysis in day 7 human blastocysts produced by in vitro fertilization. Reproductive Biology and Endocrinology. 14(1):20
Kaartinen N, Das P, Kananen K, Huhtala H, Tinkanen H (2015) Can repeated IVF–ICSI-cycles be avoided by using blastocysts developing from poor-quality cleavage stage embryos? Reproductive biomedicine online. 30(3):241–247
Wood JW (2017) Dynamics of human reproduction: biology, biometry, demography: Routledge
Maurer M, Ebner T, Puchner M, Mayer RB, Shebl O, Oppelt P et al (2015) Chromosomal aneuploidies and early embryonic developmental arrest. International journal of fertility & sterility. 9(3):346
Baart E, van Opstal D. Chromosomes in early human embryo development: incidence of chromosomal abnormalities, underlying mechanisms and consequences for development. 2014.
McCoy RC (2017) Mosaicism in preimplantation human embryos: when chromosomal abnormalities are the norm. Trends in Genetics. 33(7):448–463
Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR et al The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertility and sterility, 2014 101(3):656–63. e1
Griffin DK, Ogur C (2018) Chromosomal analysis in IVF: just how useful is it? Reproduction. 156(1):F29–F50
Teoh JP, Maheshwari A (2017) Ongoing developments in ART and pregnancy outcome. Clinical Management of Pregnancies following ART: Springer:229–242
Faramarzi A, Khalili MA, Agha-Rahimi A, Omidi M (2017) Is there any correlation between oocyte polarization microscopy findings with embryo time lapse monitoring in ICSI program? Archives of Gynecology and Obstetrics. 295(6):1515–1522
Faramarzi A, Khalili MA, Ashourzadeh S (2017) Oocyte morphology and embryo morphokinetics in an intra-cytoplasmic sperm injection programme. Is there a relationship? Zygote. 25(2):190–196
Halvaei I, Khalili MA, Esfandiari N, Safari S, Talebi AR, Miglietta S et al (2016) Ultrastructure of cytoplasmic fragments in human cleavage stage embryos. Journal of assisted reproduction and genetics. 33(12):1677–1684
Saiz IC, Gatell MCP, Vargas MC, Mendive AD, Enedáguila NR, Solanes MM et al (2018) The embryology interest group: updating ASEBIR’s morphological scoring system for early embryos, morulae and blastocysts. Medicina Reproductiva y Embriología Clínica. 5(1):42–54
Vera-Rodriguez M, Chavez SL, Rubio C, Pera RAR, Simon C (2015) Prediction model for aneuploidy in early human embryo development revealed by single-cell analysis. Nature communications. 6
Hardarson T, Caisander G, Sjögren A, Hanson C, Hamberger L, Lundin K (2003) A morphological and chromosomal study of blastocysts developing from morphologically suboptimal human pre-embryos compared with control blastocysts. Human Reproduction. 18(2):399–407
Muhlmann MC, Laudicina AO, Perandones C, Bertolino MV, Marazzi A, Quintans CJ et al (2005) Uses and limitations of two molecular cytogenetic techniques for the study of arrested embryos obtained through assisted reproduction technology. Genet Mol Res. 4(2):143–151
Riegel M (2014) Human molecular cytogenetics: from cells to nucleotides. Genetics and molecular biology. 37(1):194–209
Fesahat F, Montazeri F, Sheikhha MH, Saeedi H, Firouzabadi RD, Kalantar SM (2017) Frequency of chromosomal aneuploidy in high quality embryos from young couples using preimplantation genetic screening. International Journal of Reproductive BioMedicine. 15(5):297
Chung H-D, Kang H-Y, Jeong HJ, Lee JH, Chung MK, Park S-J (2013) Comprehensive chromosome analysis of blastocysts before implantation using array CGH. Molecular cytogenetics. 6(1):22
Shaw-Jackson C, Bertrand E, Becker B, Colin J, Beaudoin-Chabot C, Rozenberg S et al (2013) Vitrification of blastocysts derived from fair to poor quality cleavage stage embryos can produce high pregnancy rates after warming. Journal of assisted reproduction and genetics. 30(8):1035–1042
Sallem A, Santulli P, Barraud-Lange V, Le Foll N, Ferreux L, Maignien C et al (2017) Extended culture of poor-quality supernumerary embryos improves ART outcomes. Journal of assisted reproduction and genetics.:1–9
McCormick S, Pospisil C, Smith R, Schoolcraft W, Katz-Jaffe M (2018) Euploid embryos that reach the blastocyst stage on day 5 of development have a significantly higher chance of implantation. Fertility and Sterility. 110(4):e348–e3e9
Harton GL, Cinnioglu C, Fiorentino F (2017) Current experience concerning mosaic embryos diagnosed during preimplantation genetic screening. Fertility and sterility. 107(5):1113–1119
Taylor TH, Gitlin SA, Patrick JL, Crain JL, Wilson JM, Griffin DK (2014) The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Human reproduction update. 20(4):571–581
Vera-Rodriguez M, Rubio C (2017) Assessing the true incidence of mosaicism in preimplantation embryos. Fertility and sterility. 107(5):1107–1112
Munné S (2018) Status of preimplantation genetic testing and embryo selection. Reproductive BioMedicine Online.
Capalbo A, Rienzi L (2017) Mosaicism between trophectoderm and inner cell mass. Fertility and sterility. 107(5):1098–1106
Liñán A, Lawrenz B, El Khatib I, Bayram A, Arnanz A, Rubio C et al (2018) Clinical reassessment of human embryo ploidy status between cleavage and blastocyst stage by next generation sequencing. PloS one. 13(8):e0201652
Munné S, Wells D (2017) Detection of mosaicism at blastocyst stage with the use of high-resolution next-generation sequencing. Fertility and Sterility
Inoue N, Lopez R, Delgado A, Nuñez D, Portella J, Noriega-Hoces L et al (2017) Mosaic embryo transfer after oocyte in vitro maturation in combination with non-invasive prenatal testing (NIPT)—first report of a euploid live birth. Journal of Assisted Reproduction and Genetics. 34(9):1199–1205
Greco E, Minasi MG, Fiorentino F (2015) Healthy babies after intrauterine transfer of mosaic aneuploid blastocysts. New England Journal of Medicine. 373(21):2089–2090
Gleicher N, Vidali A, Braverman J, Kushnir VA, Barad DH, Hudson C et al (2016) Accuracy of preimplantation genetic screening (PGS) is compromised by degree of mosaicism of human embryos. Reproductive Biology and Endocrinology. 14(1):54
Marin D, Scott RT Jr, Treff NR (2017) Preimplantation embryonic mosaicism: origin, consequences and the reliability of comprehensive chromosome screening. Current Opinion in Obstetrics and Gynecology. 29(3):168–174
Esfandiari N, Bunnell ME, Casper RF (2016) Human embryo mosaicism: did we drop the ball on chromosomal testing? Journal of assisted reproduction and genetics. 33(11):1439–1444
Huang J, Zhao N, Wang X, Qiao J, Liu P (2015) Chromosomal characteristics at cleavage and blastocyst stages from the same embryos. Journal of assisted reproduction and genetics. 32(5):781–787
Adler A, Lee H-L, McCulloh DH, Ampeloquio E, Clarke-Williams M, Wertz BH et al (2014) Blastocyst culture selects for euploid embryos: comparison of blastomere and trophectoderm biopsies. Reproductive biomedicine online. 28(4):485–491
Acknowledgements
The authors would like to thank the colleagues who helped with the experiments and data collection. This study was supported by research and clinical center for infertility, Shahid Sadoughi University of Medical Sciences.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Conception and design of study: Fatemeh Taheri and Mohammad Ali Khalili; acquisition of data: F. Taheri, Seyed Mehdi Kalantar, Farzaneh Fesahat, and Fatemeh Montazeri; analysis and/or interpretation of data: F. Taheri, F. Fesahat, and F. Montazeri; drafting the manuscript: F. Taheri and M A. Khalili; and revising the manuscript critically for important intellectual content: M A. Khalili, M.G. Palmerini, and Bryan Woodward (PhD). All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Research and Clinical Institute for Infertility in Yazd. The study was approved by the institute ethics committee (IR.SSU.RSI.REC.1394.15).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Taheri, F., Khalili, M.A., Kalantar, S.M. et al. Generation of viable blastocysts from discarded human cleavage embryos. Middle East Fertil Soc J 25, 18 (2020). https://doi.org/10.1186/s43043-020-00028-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43043-020-00028-1