Abstract
Background
Antimicrobial peptides were examined as an evolutionarily preserved component of the innate immune response and identified as a vital first-line defense against a broad spectrum of pathogens in fish. The objective of this study was to investigate the molecular characterization and expression analysis of hepc1 and hepc2 in three strains of tilapia species infected naturally in Lake Manzala.
Results
The results revealed that the alignment of nucleotide sequences including cDNA and deduced amino acid sequences showed that hepcidin 1 in Sarotherodon galilaeus has four genotypes due to SNPs in codon 34Gln (CAG)/Leu (CTG) and codon 36Glu (GAA/GAG). Regarding hepcidin 2 gene, different genotypes were detected in Tilapia zillii and Sarotherodon galilaeus due to SNPs in codons 19Met (ATG)/Ile (ATT), 57Pro (CCA)/Ser (TCA), and codon 14Leu (CTT / CTC). Hepcidins 1 and 2 coding region sequences in three tilapia species deposited to GeneBank and phylogenetic analysis indicated that tilapia species are more similar to each other and closely related to Sea perch. On the other hand, the expression levels of hepc1 and hepc2 genes were over-expressed in different tilapia tissue species (hepc1 in Sarotherodon galilaeus and hepc2 in Tilapia zillii).
Conclusions
The results concluded that the hepcidin 1 and 2 genes showed constitutive expressions in most of tested tissues and have a very similar three-dimensional structure as well as mature peptides which mean that these genes are highly conserved within the species examined.
Similar content being viewed by others
Introduction
Fish have developed several defense mechanisms against waterborne pathogens with one of the first lines of defense consisting relatively small peptides called antimicrobial peptides (AMPs) (Neves et al. 2015). Antimicrobial peptides are identified as a positively charged molecules with short amino acid chains which play crucial roles in host defense against pathogens through antimicrobial actions or immunomodulatory functions (Jenssen et al. 2006). Among these peptides, a cystine-rich molecule isolated from human urine named hepcidin (Hep) is based on its hepatic expression and antibacterial effect in vitro (Falzacappa and Muckenthaler 2005). Hepcidin has a dual function as a key regulator of iron metabolism and as a defense peptide (Ganz 2003). Fish hepcidin was first identified and isolated from the hybrid striped bass (Hirono et al. 2005), and since then, hepcidins have been identified in at least 37 fish species (Masso-Silva and Diamond 2014). In fish and mammals, hepcidin genes consist of three exons and two introns which encode a pre-pro peptide, an acidic propiece, and a mature peptide (Douglas et al. 2003). The pre-pro peptides contain a furin cleavage site, and the mature peptide contains eight conserved cysteines and a conserved glycine residue (Asmamaw 2016).
Fish hepcidin are highly expressed in the liver and kidney while they are less expressed in the heart, brain, skin, intestine, blood cells, spleen, and stomach and unnoticeable in muscle (Asmamaw 2016). Earlier studies found that the expression of hepcidin is upregulated during infection, inflammation, and iron overload and predominantly reduced under certain conditions like anemia and hypoxia (Ganz 2007). The development of hepcidins are assorted in teleost fishes as it is influenced by the diversity of aquatic environments in which it is found (Lee et al. 2012). It has been found that the two hepcidin peptides identified in teleost fish show a degree of sub functionalization of its functions, where hepcidin 1 (HAMP1) more implicated in iron metabolism regulation and hepcidin 2 (HAMP2) mainly performing an antimicrobial role (Neves et al. 2015). The AMP production go through a main pathway by the recognition of pathogen-associated molecular patterns (PAMPs) through receptors (PRRs), such as Toll-like receptors (TLRs), exciting a cascade of intracellular signaling involving MyD88, TRAF6, IRAK1, and IKK that promotes the activation and translocation of NF-kB to the nucleus where it transcriptionally stimulates AMPs and other effector molecules (Campoverdea et al. 2017).
In fact, the structures and sequences of hepcidin genes are highly preserved between mammals and fish (Hsieh et al. 2010). According to previous studies on different fish species, two types of hepcidins have been identified in turbot (Scophthalmus maximus), using Matras with approximately 50% sequence identity; the synthesized form of these peptides showed apparent bactericidal activities against both Gram-positive and Gram-negative bacteria in a dose-dependent manner (Jain et al. 2015). Concerning the analysis results of genomic characterization and gene expression of four hepcidin genes in the red-banded seabream (Pagrus auriga), it was apparent that although hepcidin antimicrobial peptides (HAMP) might play an important role against bacterial infections, it also has additional functions in iron regulation and embryogenesis in fish (Martin-Antonio et al. 2009). Former study by Pridgeon (2012) demonstrated relative transcriptional levels of AMHP, using quantitative polymerase chain reaction (QPCR), in response to Edwardsiella ictaluri infection. After identification and characterization of AMHP in miiuy croaker, Xu et al. (2012) concluded that the expression level of these peptides was highest in the liver and an upregulation of transcription was seen in the intestine, spleen, and kidney in response to pathogenic bacteria, Vibrio anguillarum, which implied that HAMP has a vital role as a first line of defense against invading pathogens.
The objective of this study was to investigate the molecular characterization and expression analysis of hepc1 and hepc2 in three strains of tilapia species infected naturally in Lake Manzala.
Materials and methods
One hundred and ten fishes were collected from Lake Manzala in Egypt.
- a.
Collection of samples
Tissue samples were obtained from normal and naturally infected fish in Lake Manzala. The selected tissues were gonads, spleen, head kidneys, and liver. The fish was kept on ice, while the tissues were kept in Hank’s Balanced Salt Solution (HBSS), then they were homogenized and the PeqGold TriFast™ (or Trisure or Tripure) (stabilization solution) was added. After this addition, the RNA was not degraded by Rnase and the samples could be stored at − 80 °C indefinitely. Individual fish were dissected, and these samples were collected for bacteriological examination, and isolation of some types of bacteria was attempted according to Noga (1996).
- b.
RNA isolation and cDNA synthesis
Total RNA was isolated from several tissues (gills, kidney, liver, and spleen) of Oreochromis niloticus, Sarotherodongalilaeus, and Tilapia zillii.Tissue samples were separately ground quickly in liquid nitrogen and transferred to 1 mL TRIzol solution (cat#15596-026, Invitrogen, Germany) to isolate RNA following the manufacturer’s instructions. RNA was quantified and checked for purity using a NanoDrop (Thermo Scientific, Waltham, MA, USA). Integrity was assured with ethidium bromide-stain analysis of 28S and 18S bands by formaldehyde-containing agarose gel electrophoresis. Extracted RNA was stored in a − 80 °C freezer until used as template for reverse transcription PCR (RT-PCR). One microliter of RNA isolated from several tissues was reverse transcribed into cDNA in a total volume of 20 μl using Revert AidTM First Strand cDNA Synthesis Kit (MBI Fermentas, Germany) according to the manufacturer’s instructions.
- c.
Primer design and PCR optimization
The specific primers for the antimicrobial peptide genes were designed using known sequences of Teleost AMPs using the methodology of the homology cloning. Eleven pairs of primers were designed using the free primer3 online software (http://frodo.wi.mit.edu/primer3/), based on the NCBI reference sequences. Accession numbers of the genes for which the primers are designed are listed in Table 1. They are aimed to amplify fish antimicrobial peptides messenger RNAs (mRNAs) using PCR. All the primers pairs were designed to amplify full coding sequence of the target antimicrobial peptide mRNA. The designed primers to amplify hepcidin mRNA in Oreochromis niloticus, Tilapia zillii, and Sarotherodon galilaeus based on the conserved region in hepcidin mRNA of bony fish. The primers were synthesized by HVD life sciences GMBH, Germany. Designed primers were tested in order to determine its optimum annealing temperature that amplifies the target cDNA segment.
- d.
Polymerase chain reaction (PCR) amplification
PCR amplification of cDNA was performed according to the method described by Ausubel et al. (1990). The optimized PCR cycles were differed according to the gene under study. The PCR cycle started with initial denaturation of 94 °C for 5 min, then followed by 35 cycles (three steps) as: 94 °C for 30 s, annealing temperature for 45 s. with variable temperatures according to the gene under study, then extension at 72 °C for 1 min and finally one step for final extension 72 °C for 5 min. The annealing temperatures were as follows: hepcidin 1 (53 °C) and hepcidin 2 (55 °C). For the PCR optimization and detection of the PCR success, some of the PCR products were subject to run horizontally on agarose electrophoresis and stained with ethidium bromide.
- e.
Sequencing, alignment analysis and amino acid translation
Prior to sending the PCR products to the company for sequencing, the PCR products were purified using GeneJET Gel Extraction Kit from Thermo scientific Company according to the manufacturer’s instructions. PCR products from two genes (hepcidin 1 and hepcidin 2) of three tilapia fish species were sent for sequencing in Macrogen Company (South Korea).
Statistical analysis
Sequence analysis was performed by multiple alignments using CLUSTAL-W program (Thompson et al. 1994). Protein translations of CDs sequences were carried out using open reading frame finder (https://www.ncbi.nlm.nih.gov/orffinder/) while modeling of hepcidin protein was determined by Phyre2 software (http://www.sbg.bio.ic.ac.uk/phyre2/). For the analysis of potential cleavage site of the signal peptide, SignalP Software was used (http://www.cbs.dtu.dk/services/SignalP/). The alignment of the amino acid sequences of hepcidin proteins was done using Clustal X. The phylogenetic tree was drawn by the neighbor-joining method (Saitou and Nei 1987) and analyzed with MEGA-3 (Kumar et al. 2018).
Quantitative evaluation of hepcidin 1 and 2 mRNA by real-time PCR (qPCR)
StepOne™ Real-Time PCR System from Applied Biosystems (Thermo Fisher Scientific, Waltham, MA USA) was used to determine the tilapia’s cDNA copy number. Sequencing results of different hepcidin genes were used to design gene specific primers, for qPCR, by using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/ primer3_www.cgi). β-Actin gene used as internal control to correct for differences in total cDNA input between samples. PCR reactions were set up in 25 μL reaction mixtures containing 12.5 μL × 1 SYBR® Premix Ex TaqTM (TaKaRa, Biotech. Co. Ltd.), 0.5 μL 0.2 μM sense primer, 0.5 μL 0.2 μM antisense primer, 6.5 μL distilled water, and 5 μL of cDNA template. The qPCR conditions were 95.0 °C for 10 min followed by 40 cycles of 95.0 °C for 15 s and 60.0 C for 1 min. The relative amount of the genes of interest was calculated by the comparative threshold cycle (CT) method with efficiency correction, using the mean PCR efficiency for each amplicon (Ruijter et al. 2009). The sequences of QPCR primers are listed in Table 1.
Results
Naturally infected tilapia
Three strains of fish including O. niloticus, T. zillii, and S. galilaeus were naturally infected with bacteria in the Lake Manzala. The O. niloticus strain was infected with Aeromonas hydrophila but T. zillii and S. galilaeus were infected by Pseudomonas luteola.
Polymerase chain reaction (PCR)
Hepcidin 1 gene-specific primers amplified a product of 264 bp and hepcidin 2 gene-specific primers amplified a product of 273 bp (Fig. 1).
Characterization of hepcidin 1 coding region sequences in O. niloticus, T. zillii, and S. galilaeus
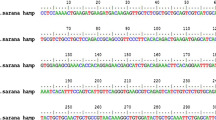
The O. niloticus, T. zillii, and S. galilaeus hepcidin 1 coding region (CDs) were sequenced, and their sequences have been deposited to GeneBank with accession numbers of (MH651353, MH651354, and MH651358), respectively. CD regions of T. zillii and S. galilaeus have the same length (264 bp) while O. niloticus has 258-bp CD length with a deletion portion (AGCAGG) when aligned with T. zillii and S. galilaeus CDs (Fig. 2). Moreover, two SNPs are determined among these species at nt 36 (C/G) and at nt 240 (A/C). Four genotypes are determined in S. galilaeus species due to the presence of SNPs at nt 101 W(A/T) and nt 108 R (G/A) while no different genotypes are detected in O. niloticus and T. zillii species (Fig. 2).
The deduced amino acid sequences for hepcidin 1 CDs of T. zillii and S. galilaeus consists of 87 amino acids. One SNP at nt 101 which presents in codon 34 (CAG/CTG) in S. galilaeus species is translated glutamine (Q) into leucine (L) while the other SNP at nt 108 which correspond to codon 36 (GAA/GAG) is silent mutation as in Fig. 2a, b. On the other hand, the amino acid sequence for hepcidin 1 CD of O. niloticus species is 85 amino acids due to deletion portion which detected in nucleotide sequence as shown in Fig. 3 a, b and c.
Sequence alignment and phylogenetic analysis
The deduced amino acid sequences of hepcidin 1 in O. niloticus, T. zillii, and S.galilaeus have been aligned with protein sequences of turbot_S.maximus (AY994074), Homo sapiens (BC020612), white bass (AAM28440), Atlantic salmon (BI468191), Canis lupus familiaris (AY899807), mus musculus (BC021587), black seabream (AAU00801), red seabream (AAR28076), Pig (NP_999282), sea perch (AAT09138), Japanese flounder (BAE06234), winter flounder (AW013026), channel catfish (AAX39713), blue catfish (AAX39714), and zebrafish (P61516) as in Fig. 4. It is obvious that O. niloticus, T. zillii, and S. galilaeus hepcidin 1 gene is more similar to each other and closely related to sea perch than other species (Figs. 4 and 5).
Alignment of deduced amino acid sequences of O. niloticus, T. zillii, and S. galilaeus hepcidin 1 gene with those of turbot_S.maximus (AY994074), Homo sapiens (BC020612), white bass (AAM28440), Atlantic salmon (BI468191), Canis lupus familiaris (AY899807), Mus musculus (BC021587), black seabream (AAU00801), red seabream (AAR28076), pig (NP_999282), sea perch (AAT09138), Japanese flounder (BAE06234), winter flounder (AW013026), channel catfish (AAX39713), blue catfish (AAX39714), and zebrafish (P61516)
Phylogenetic analysis of O. niloticus, T. zillii, and S. galilaeus hepcidin 1 gene and other species. Constructions are formed on the basis of the homology sequences calculated from the complete amino acid sequence of hepcidin genes. The tree is constructed using a neighbor-joining method. The scale is 0.20 which refers to percentage of divergence
Characterization of hepcidin 2 coding region sequences in O. niloticus, T. zillii, and S. galilaeus
The O. niloticus, T. zillii, and S. galilaeus hepcidin 2 CDs were sequenced, and their sequences have been deposited to GeneBank with accession numbers of (MH651359.1, MH651363.1, and MH651367.1), respectively. CD regions in these species have the same length (273 bp) with identical sequence except in two SNPs at positions nt 252 (T/C) and nt 310 (G/A). Four genotypes are determined in T. zillii due to the presence of SNPs at nt 101 K(G/T) and nt 213 Y(C/T). In addition, four genotypes are detected in S. galilaeus due to the presence of SNPs at nt 86 Y(C/T) and nt 101 K(G/T) while no different genotypes are detected in O. niloticus (Fig. 6).
cDNA sequence of hepcidin 2 gene. The coding regions start codon ATG and termination codon TGA are both bold and underlined. The alignment of nucleotide sequences of O. niloticus, T. zillii, and S. galilaeus shows two SNPs at nt 252 (T/C) and nt 310 (G/A) (in red box) and different genotypes for each species (in blue box)
The deduced amino acid sequence for hepcidin 2 gene of O. niloticus, T. zillii, and S. galilaeus consists of 90 amino acids. The two SNPs at nt 101 and nt 213 are present in codons 19 (ATG/ATT) and 57 (CCA/TCA) in T. zillii species are translated methionine (M) into isoleucine (I) and proline (P) into Serine (S) as shown in Fig. 7a, b. On the other hand, S. galilaeus species has SNP at nt 101 which present in codon 19 (ATG/ATT) is translated into methionine (M) into isoleucine while, the other SNP at nt 86 which correspond to codon 14 (CTT/CTC) is a silent mutation as in Fig. 7c, d. In addition, the only genotype detected for O. niloticus has methionine (M) at codon 19 (ATG) as in Fig. 7e.
Sequence alignment and phylogenetic analysis
The deduced amino acid sequences of hepcidin 2 in O. niloticus, T. zillii, and S. galilaeus have been aligned with protein sequences of turbot_S.maximus (AY994074), Homo sapiens (BC0 C0 20612), white bass (AAM28440), Atlantic salmon (BI468191), Canis lupus familiaris (AY 89980897), Mus musculus (BC021587), black seabream (AAU00801), red seabream (AAR28076), pig (N P_ P_999282), sea perch (AAT09138), Japanese flounder (BAE06234), winter flounder (AW W1013026), channel catfish (AAX39713), Blue catfish (AAX39714), zebrafish (P61516) as in Fig. 8. It is obvious that O. niloticus, T. zillii, and S. galilaeus hepcidin 2 gene is more similar to each other and closely related to sea perch than other species (Fig. 9).
Alignment of deduced amino acid sequences of O. niloticus, T. zillii, and S. galilaeus hepcidin 2 gene with those of turbot_S.maximus (AY994074), Homo sapiens (BC020612), white bass (AAM28440), Atlantic salmon (BI468191), Canis lupus familiaris (AY899807), mus musculus (BC021587), black seabream (AAU00801), red seabream (AAR28076), pig (NP_999282), sea perch (AAT09138), Japanese flounder (BAE06234), winter flounder (AW013026), channel catfish(AAX39713), blue catfish (AAX39714), and zebrafish (P61516)
Phylogenetic analysis of O. niloticus, T. zillii, S. galilaeus hepcidin 2 gene and other species. Constructions are formed on the basis of the homology sequences calculated from the complete amino acid sequence of hepcidin genes. The tree is constructed using a neighbor-joining method. The scale is 0.20 which refers to percentage of divergence
Structural model analysis of hepcidin 1 and hepcidin 2 and signal peptide detection
Three-dimensional structure of hepcidin 1 and hepcidin 2 shows it to be an amphipathic molecule composed of two distorted antiparallel β-sheets separated by a hairpin loop, containing a vicinal disulfide bond (disulfide bond between adjacent cysteines), and stabilized by three inter-β-sheet disulfide bonds which remarkably similar among O. niloticus, T. zillii, and S. galilaeus species as shown in Fig. 10a and b. On the other hand, the signal peptide is detected from 1 to 24 amino acids with high probability (0.9753 and 0.9968, respectively) and mature protein starts at position 25 amino acid (Fig. 11a, b).
Gene expression results
Expression of antimicrobial peptide gene hepcidin 1 (H1) in O. niloticus, T. zillii, and S. galilaeus naturally infected with bacteria in the Lake Manzala
The expression levels of the antimicrobial peptide gene hepcidin 1 (H1) in several tissues of O. niloticus, T. zillii, and S. galilaeus were quantified by real-time RT-PCR (Fig. 12). The results revealed that expression levels of h1 gene were increased in tissues of S. galilaeus compared with those of O. niloticus and T. zillii. Moreover, the lowest expression levels of h1 gene were observed in tissues of the T. zillii. Additionally, the highest expression levels were observed in the gills and spleen tissues of all fish strains compared with kidney and liver tissues.
Expression of antimicrobial peptide gene hepcidin 2 (H2) in O. niloticus, T. zillii, and S. galilaeus naturally infected with bacteria in the Lake Manzala
The expression levels of the antimicrobial peptide gene hepcidin 2 (H2) in several tissues of O. niloticus, T. zillii, and S. galilaeus were quantified by real-time RT-PCR (Fig. 13). The results revealed that expression levels of H2 gene were increased in tissues of T. zillii compared with those of O. niloticus and S. galilaeus. Moreover, the lowest expression levels of H2 gene were observed in tissues of the O. niloticus. Furthermore, the highest expression levels were observed in the kidney and liver tissues of all fish strains compared with gills and spleen tissues.
Discussion
Fish is protecting their biological systems and fighting toward many of pathogens by producing a several types of AMPs (Rajanbabu and Chen 2011). In our knowledge, there are no published data concerning AMPs in S. galilaeus and T. zillii. The results of the present study found that sequence alignment for hepcidin 1 and 2 CDs of S. galilaeus and T. zillii have the same length; the sequence identity is 99.24% while the CD identity between S. galilaeus and O. niloticus is 100%. The results showed that S. galilaeus possessed four genotypes in hepcidin 1 due to SNPs in codon 34Gln (CAG)/Leu (CTG) and codon 36Glu (GAA/GAG) (silent mutation) and different genotypes in hepcidin 2 due to SNPs in codon 19Met (ATG)/Ile (ATT) and codon 14Leu (CTT/CTC) (silent mutation). On the other hand, different genotypes recorded in hepcidin 2 gene of T. zillii due to SNPs in codons 19Met(ATG)/Ile (ATT) and 57Pro (CCA)/Ser (TCA). Previous studies stated that two forms of hepcidins were identified in turbot (Scophthalmus maximus) with 50% identity of sequencing (Zhang et al. 2014). In Miiuy croaker, two hepc1 and 2 genes were identified with sequence similarity 99.6% (Xu et al. 2012). The sequence alignment of putative hepcidin genes in Nile and blue tilapia fishes, hepcidin 1 and hepcidin 2 represented identity > 50% and > 80% (Bian et al. 2019). Parallel to our results, a study by Bao et al. (2006) reported that the sequence alignment of channel and blue catfish hepc2 differ in two amino acids at two positions: (a) at 28, the channel catfish hepc2 has a proline residue while the blue catfish hepc2 has a serine residue, (b) at 32, the channel catfish hepc2 has an arginine residue while the Blue catfish hepc2 has a proline residue. The two hepc2 cDNAs of channel and blue catfish had 25 single-nucleotide polymorphisms. So far, Neves et al. (2015) detected high number of SNPs in the sequence of mature peptide (~ 7 SNPs) compared with the remainder of prepropeptide (only 1 SNP) of hamp2 gene in sea bass. Furthermore, alignment of the two types of hepcidin in sea bass exhibited differences in the amino acids between both gene sequences of Hamp1 and Hamp2. However, the sequence of the mature peptides exhibited unchanged eight cysteine residues sequences at the conserved positions.
The action mechanism of how sequence differences in the SNPs formation is that it could be possible during life generation of the fish, various type of deletion or insertion might be occurred in the introns of the hepcidin sequences. However, molecular analysis in fish exhibited that a slight differences are occurred because the sequences of hamp1 and hamp2 genes are remain conserved.
The deduced amino acid sequences for hepcidin 1 CDs of T. zillii and S. galilaeus consists of 87 amino acids for O. niloticus 85 aa due to deletion portion in mature peptide while those for hepcidin 2 CDs encodes a peptide of 90aa in three tilapia species. Previous work by Bao et al. (2006) reported that hepcidin 2 of the blue catfish and channel catfish shared similar features as it encodes a peptide of 94 aa. The results of numerous studies conducted by Wang et al. (2012), Hu et al. (2007), and Shi and Camus (2005) showed that a hepcidin of 78 aa and 96aa were detected in Tor putitora, Ictalurus punctatus, Cynoglossus semilaevis, respectively. As well as, results by Neves et al. (2015) detected that hepc1 and hepc2 genes of teleost fish encode 91aa and 85aa, respectively. The miiuy croaker HAMP was predicted to encode of 89 amino acids (Xu et al. 2012) which in consistent with results of Silva et al. (2011), where A. immaculatus prepro hepcidin is composed of 90 aa. It was concluded that the conserved functions of fish hepcidin is due to the conservation of their mature peptide.
Phylogenetic analysis indicated that translated protein of hepcidins CDs of the species examined are organized as clusters and very related to sea perch while so far from two or more hepcidins of winter flounder, Atlantic salmon, and mouse. Lauth et al. (2005) had shown that fish hepcidins in bass and tilapia species were characterized by cysteine-rich peptides and a central model of four conserved disulfide bridges. In addition, a study by Huang et al. (2007) revealed that tilapia hepcidin (2–3) situated in a group with Japanese flounder hepcidin-JF1, 2 and tilapia hepcidin 2–2 while tilapia hepcidin1–5 is closest to white bass and black porgy which suggest that the three tilapia hepcidins may have different functions. Based on results recorded by Silva et al. (2011), HAMP-1of A. Immaculatus had a sequence similarity of 96% with Serranid fish and Epinephelus moara and grouped with A. immaculatus in the same cluster. According to results reported by Neves et al. (2015), phylogenetic analysis cluster fish hepcidins 1& 2 of Perciformes and Pleuronectiformes together separated from other vertebrate hepcidins. Additionally, Campoverde et al. (2017) found that the phylogenetic analysis revealed that meager hep2 sequence was more similar to that of Larimichthys crocea. Even though, there are antithesis opinions suggesting that progression and divergence of hepcidin antimicrobial peptide is connected with their surrounding habitats and pathogens.
This study reported that the three-dimensional structure of hepcidin 1 and hepcidin 2 shows it to be an amphipathic molecule composed of two distorted antiparallel β-sheets separated by a hairpin loop, containing a vicinal disulfide bond (disulfide bond between adjacent cysteines), and stabilized by three inter-β-sheet disulfide bonds and signal peptide is detected from 1 to 4 amino acids which remarkably similar among O. niloticus, T. zillii, and S. galilaeus species. Xu et al. (2008) demonstrated that lower vertebrates have very similar structures of hepcidin homologs (20-25aa) with eight highly conserved cysteines and four intramolecular disulfide bonds, giving hepcidin a hairpin structure. Tilapia fish Oreochromis mossambicus (Huang et al. 2007) and schizothorax richardsonii (Gray) (Chaturvedi et al. 2014) presented the same results. Previous studies by Zhang et al. (2009), Wang et al. (2010), and Chen et al. (2009) demonstrated that hepcidin hairpin structure with cationic residual act against fungi, bacteria, tumor cells, and viruses. Hepcidin hairpin-like structure are considered to be essential for its activity (Neves et al. 2015).
Regarding gene expression results, this study found that expression levels of H1 and H2 genes were increased in gills, kidney, and liver tissues of O. niloticus, T. zillii, and S. galilaeus naturally infected with bacteria in the Lake Manzala. However, the expression levels of H1 and H2 genes were decreased in spleen tissues. In the same line with our findings, several studies found that in many fish species and other vertebrates (Krause et al. 2000; Park et al. 2001; Zhou et al. 2011; Li et al. 2013; Chen et al. 2007; Rodrigues et al. 2006), both sea bass H1 and H2 genes were abundantly expressed in the liver. Additionally, they also reported that the expression levels of H1 and H2 genes were very low in the spleen. Hepcidin expression has been reported to have an important role in macrophages, lymphocytes, and monocytes but further analysis will be required to determine the exact cells responsible for this expression (Costa et al. 2011; Sow et al. 2007; Zhang and Rovin 2010; Pinto et al. 2010). The higher antimicrobial potential of H1 and H2 genes were observed in this study with over-expression in gills. That could be explained because these tissues are in constant contact with the surrounding environment, many times a doorway to microbial infection.
The present study found that strain of fish including O. niloticus, T. zillii, and S. galilaeus were naturally infected with bacteria in the Lake Manzala. The O. niloticus strain was infected with Aeromonas hydrophila but T. zillii and S. galilaeus were infected by Pseudomonas luteola. Several studies reported that both H1 and H2 genes were upregulated in fish with bacterial infections (Hirono et al. 2005; Huang et al. 2007). A significant induction of both sea bass H1 and H2 gene types was observed after experimental bacterial challenge with P. damselae spp. piscicida.
Additionally, it has been reported that H1 and H2 genes may have antimicrobial activity even so the initial pathogen density is very low (4 3 103 CFU/ml) (Zhang et al. 2014). Moreover, H1 and H2 peptides manifested strong antimicrobial activities against Gram-positive bacteria. Further studies will be required to better understand the relevance of each type of hepcidin genes and to gain insight into their antimicrobial properties.
Conclusions
The results concluded that the hepcidin 1 and 2 genes showed constitutive expressions in most of tested tissues and have a very similar three-dimensional structure as well as mature peptides which mean that these genes are highly conserved within the species examined.
Availability of data and materials
All data generated or analyzed during this study are included in this published article
Change history
16 April 2020
After publication of the original article [1], we were notified that Figure 1 is incorrect. The corresponding author uploaded the wrong figure by mistake.
Abbreviations
- Hepc1:
-
Hepcidin 1
- Hepc2:
-
Hepcidin 2
- T. zillii :
-
Tilapia zillii
- S. galilaeus :
-
Sarotherodon galilaeus
- O. niloticus :
-
Oreochromis niloticus
- HAMP1:
-
Hepcidin Antimicrobial peptide 1
- HAMP2:
-
Hepcidin Antimicrobial peptide 2
- cDNA:
-
Complementary DNA
- AMPs:
-
Antimicrobial peptides
- β-actin:
-
Beta actin
- CD:
-
Coding region
- aa:
-
Amino acid
- mRNA:
-
Messenger RNA
- RT-PCR:
-
Real time –polymerase chain reaction
References
Asmamaw B (2016) Hepcidin and Its Roles in Fishes. The Journal of Zoology Studies. 3(3):1–10
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Kevin S. Current Protocols in Molecular Biology In: the polymerase chain reaction K. B. Mullis, F. Ferre, R.A. Gibbsditors. 1990 (eds.)
Bao B, Peatmana E, Xua P, Lia P, Zenga H, Hea C, Liua BZ (2006) The catfish liver-expressed antimicrobial peptide2 (LEAP-2) gene is expressed in a wide range of tissues and developmentally regulated Baolong. Mol Immunol 43:367–377
Bian C, Li J, Lin X, Chen X, Yi Y, You X, Zhang Y, Lv Y, Shi Q (2019) Whole genome sequencing of the blue tilapia (Oreochromis aureus) provides a valuable genetic resource for biomedical research on tilapias. Mar Drugs. 17:386
Campoverdea C, Douglas J, Milne B, Est A, Evez A, Neil D, Christopher JS, Karl BA (2017) Ontogeny and modulation after PAMPs stimulation of b-defensin, hepcidin, and piscidin antimicrobial peptides in meagre (Argyrosomu sregius). Fish & Shellfish Immunology. 69:200–210
Chaturvedi PX, Dhanik M, Pande A (2014) Characterization and structural analysis of hepcidin like antimicrobial peptide from schizothorax richardsonii (Gray). Protein J. 33:1–10
Chen JY, Lin WJ, Lin TL (2009) A fish antimicrobial peptide, tilapia hepcidin TH2-3, shows potent antitumor activity against human fibrosarcoma cells. Peptides. 30:1636–1642
Chen SL, Li W, Meng L, Sha ZX, Wang ZJ, Ren GC (2007) Molecular cloning and expression analysis of a hepcidin antimicrobial peptide gene from turbot (Scophthalmus maximus). Fish Shellfish Immunology. 22:172–181
Costa MM, Maehr T, Diaz-Rosales P, Secombes CJ, Wang T (2011) Bioactivity studies of rainbow trout (Oncorhynchus mykiss) interleukin-6: effects on macrophage growth and antimicrobial peptide gene expression. Molecular Immunology. 48:1903–1916
Douglas SE, Gallant JW, Liebscher RS, Dacanay A, Tsoi SCM (2003) Identification and expression analysis of hepcidin-like antimicrobial peptides in bony fish. Developmental Comparative Immunol. 27:589–601
Falzacappa MVV, Muckenthaler MU (2005) Hepcidin: Iron-hormone and anti-microbial peptide. Gene. 364:37–44. https://doi.org/10.1016/j.gene..07.020
Ganz T (2003) Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood. 102(3):783–788
Ganz T (2007) Molecular control of iron transport. Journal of the American Society of Nephrology. 18:394–400
Hirono I, Hwang JY, Ono Y, Kurobe T, Ohira T, Nozaki R, Aoki T (2005) Two different types of hepcidins from the Japanese flounder Paralichthys olivaceus. Fedration Eurobean Biochemical Societies J. 272:5257–5264
Hsieh JC, Pan CY, Chen JY (2010) Tilapia hepcidin (TH)2-3 as a transgene in transgenic fish enhances resistance to Vibrio vulnificus infection and causes variations in immune-related genes after infection by different bacterial species. Fish & Shellfish Immunology. 29:430–439
Hu X, Camus AC, Aono S, Morrison EE, Dennis J, Nusbaum KE et al (2007) Channel catfish hepcidin expression in infection and anemia. Comparative Immunology, Microbiolology of Infectious Diseases. 30:55–69. https://doi.org/10.1016/j.cimid.2006.10.004
Huang PH, Chen JY, Kuo CM (2007) Three different hepcidins from tilapia, Oreochromis mossambicus: Analysis of their expressions and biological functions. Molecular Immunology. 44(8):1922–1934
Jain A, Yadav BK, Chugh A (2015) Marine antimicrobial peptide tachyplesin as an efficient nanocarrier for macromolecule delivery in plant and mammalian cells. FEBS J. 282:732–745
Jenssen H, Hamill P, Hancock REW (2006) Peptide antimicrobial agents. Clin Microbiol Rev. 19(3):491–451
Krause AS, Neitz HJ, Mägert A, Schulz WG, Forssmann P, Schulz-Knappe, Adermann K (2000) LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett 480:147–150
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35(6):1547–1549
Lauth X, Babon JJ, Stannard JA, Singh S, Nizet V, Carlberg JM et al (2005) Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J. Biological Chem. 280(10):9272–9282
Lee J, Wan K, Adnan A (2012) Molecular characterization of hepcidin in the Asian seabass (Lates calcarifer) provides insights into its role in innate immune response. Aquaculture. 330:8–14
Li H, Zhang F, Guo H, Zhu Y, Yuan J, Yang G, An L (2013) Molecular charac- terization of hepcidin gene in common carp (Cyprinus carpio L.) and its expression pattern responding to bacterial challenge. Fish Shellfish Immunol. 35:1030–1038
Martin-Antonio B, Jiménez-Cantizano RM, Salas-Leiton E, Infante C, Manchado M (2009) Genomic characterization and gene expression analysis of four hepcidin genes in the Red banded seabream (Pagrus auriga). Fish Shellfish Immunol. 26(3):483–491
Masso-Silva JA, Diamond G (2014) Antimicrobial peptides from fish. Pharmaceuticals (Basel). 7(3):265–310. https://doi.org/10.3390/ph7030265
Neves JV, Caldas C, Vieira I, Ramos MF, Rodrigues PN (2015) Multiple hepcidins in a teleost fish, Dicentrarchus labrax: different hepcidins for different roles. J Immunol. 195(6):2696–2709
Noga EJ (1996) Fish disease: diagnosis and treatment. Moshy-Year book, Inc, Naples, Tokyo, New York, p 294
Park CH, Valore EV, Waring AJ, Ganz T (2001) Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biogiological Chem. 276:7806–7810
Pinto JP, Dias V, Zoller H, Porto G, Carmo H, Carvalho F, Sousa M (2010) Hepcidin messenger RNA expression in human lymphocytes. Immunology. 130:217–230
Pridgeon JW (2012) Expression profiles of seven channel catfish antimicrobial peptides in response to Edwardsiella ictaluri infection. J Fish Dis. 35:227–237
Rajanbabu V, Chen JY (2011) Applications of antimicrobial peptides from fish and perspectives for the future. Peptides 32:415–e420. https://doi.org/10.1016/j.peptides..11.005
Rodrigues PN, Va´zquez-Dorado S, Neves JV, Wilson JM (2006) Dual function of fish hepcidin: response to experimental iron overload and bacterial infection in sea bass (Dicentrarchus labrax). Developmental. Comparative. Immunology. 30:1156–1167
Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, Hoff MJB, Moorman AFM (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:45
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4(4):406–425
Shi J, Camus AC (2005) Hepcidins in amphibians and fishes: Antimicrobial peptides or iron-regulatory hormones? Developmental Comparative Immunology 30:746–755. https://doi.org/10.1016/j.dci.10.009
Silva JM, Gill Diamond B, Maria Macias-Rodriguez A, Ascencio F (2011) Genomic organization and tissue-specific expression of hepcidin in the pacific mutton hamlet, Alphestes immaculatus (Breder, 1936). Fish Shellfish Immunol. 31:1297–1302
Sow FB, Florence WC, Satoskar AR, Schlesinger LS, Zwilling BS, Lafuse WP (2007) Expression and localization of hepcidin in macrophages: a role in host defense against tuberculosis. J Leukocytic Biol. 82:934–945
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids research. 22(22):4673–4680
Wang Y, Liu X, Ma L, Yu Y, Yu H, Mohammed S et al (2012) Identification and characterization of a hepcidin from half-smooth tongue sole Cynoglossus semilaevis. Fish Shellfish Immunol. 33:213–219. https://doi.org/10.1016/j.fsi.2012.04.011
Wang YD, Kung CW, Chen JY (2010) Antiviral activity by fish antimicrobial peptides of epinecidin-1 and hepcidin 1-5 against nervous necrosis virus in medaka. Peptides. 31(6):1026–1033
Xu Q, Cheng CH, Hu P, Ye H, Chen Z, Cao L, Chen L, Shen Y, Chen L (2008) Adaptive evolution of hepcidin genes in antarctic notothenioid fishes. Mol Biol Evol. 25:1099–1112
Xu T, Sun Y, Shi G, Wang R (2012) Miiuy croaker hepcidin gene and comparative analyses reveal evidence for positive selection. PLoS One. 7(4):35449
Zhang J, Yan Q, Ji R, Zou W, Guo G (2009) Isolation and characterization of a hepcidin peptide from the head kidney of large yellow croaker, Pseudosciaena crocea. Fish Shellfish Immunol. 26:864–870
Zhang X, Rovin BH (2010) Hepcidin expression by human monocytes in response to adhesion and pro-inflammatory cytokines. Biochemistry Biophysics Acta 1800(12):1262–1267
Zhang J, Yu LP, Li MF, Sun L (2014) Turbot (Scophthalmus maximus) hepcidin-1 and hepcidin-2 possess antimicrobial activity and promote resistance against bacterial and viral infection. Fish Shellfish Immunol. 38(1):127–134
Zhou JG, Wei JG, Xu D, Cui HC, Yan Y, Ou-Yang ZL, Huang XH, Huang YH, Qin QW (2011) Molecular cloning and characterization of two novel hepcidins from orange-spotted grouper, Epinephelus coioides. Fish Shellfish Immunol. 30:559–568
Acknowledgements
This study was financially supported by the Science and Technology Development Fund (STDF), Egypt (Grant number: 15153).
We thank Noha M. Osman who carried out the sequence data analysis and statistical analysis.
Funding
The work was funded by the Science and Technology Development Fund (STDF), Egypt (Grant number: 15153).
Author information
Authors and Affiliations
Contributions
KFM contributed her idea, designed the experiments, co-wrote the paper, and supervised the research. HAMAK performed the experiments and co-wrote the paper. DMM performed the experiments and co-wrote the paper. MMA designed the primers and performed the experiments. NMO performed the bioinformatic and statistical analyses. WKB Khalil performed the experiments and co-wrote the paper. Hassanane M. S supervised the research. All authors contributed to the study design, laboratory analysis, and writing of the manuscript. All authors revised the final manuscript and approved it.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the ethical committee of the National Research Centre
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Mahrous, K.F., Abd El - Kader, H.A.M., Mabrouk, D.M. et al. Molecular characterization and expression analysis of hepc1 and hepc2 in three tilapia species collected from Lake Manzala. Bull Natl Res Cent 44, 36 (2020). https://doi.org/10.1186/s42269-020-0278-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-020-0278-4