Abstract
Background
The phylogeny and phylogenetic relationships of Trichoderma species were investigated by many methods including maximum parsimony and distance analysis of DNA sequences from multiple genetic loci. 5.8S rDNA sequence analysis is a powerful tool in evolution studies.
Main body
The aim of this study was to identify and determine the genetic distance between 36 Trichoderma isolates from Egyptian and Saudi Arabian soils based on their 5.8S rDNA sequences. The genetic diversity of Trichoderma (Hypocreales, Ascomycota) species from Egypt and Saudi Arabia was investigated. The DNA barcoding of 36 isolates, based on the internal transcribed spacers 1 and 4 (ITS1 and 4) of the ribosomal RNA gene cluster and the partial sequence of this gene, revealed the diversity of these isolates. BLAST query showed that two species, namely T. longibarchiatum (42.7% of the isolates) and T. lentiforme (33.3% of the isolates), pre-dominated the isolates. Variation among specimens of the same species was detected. Cluster analysis of the Egyptian isolates showed two groups; the first group was subdivided into two subgroups, the first of which included T. lentiforme and T. crassum isolates, and the second subgroup harbored T. longibrachiatum, T. pseudokoningii, and T. bissettii. The second group included T. neokoningii and T. yunnanens. The cluster analysis of isolates from Saudi Arabia also consisted of two groups. The first group was subdivided into two subgroups; the first included T. longibrachiatum and T. pseudokoningii. The second subgroup harbored T. simmonsii and T. lentiforme. Also, no correlation between genetic and geographic distance was detected.
Conclusion
The results of the present study indicated that the Saudi Arabian isolates showed greater nucleotide diversity compared to Egyptian isolates. Also, these findings will assist in future studies while assessing Trichoderma genetic diversity.
Similar content being viewed by others
Background
One of the main goals of the agricultural development is to secure enough food to the growing population worldwide, where the use of biotechnology is essential in this context (Mazrou et al. 2020). The most significant agricultural-related problems in Egypt is the decrease in edible crops in the presence of an increasing population, the erosion of agricultural lands, and the loss of crops associated with diseases and pathogens (Hassan 2014 and Mazrou et al. 2020). One promising resolution to tackle these problems depends on the use of biological control. The term biological control means the use of biological materials to control plant-causing biological diseases and increase crop productivity (Hassan et al. 2019). Trichoderma spp. are of great benefit to Egyptian and Saudi Arabian agriculture. Trichoderma has many beneficial properties such as being environmental-friendly fungi, a biological control agent, and a promoter of plant growth. Several Trichoderma species (e.g., T. bissettii, T. crassum, T. lentiforme, T. longibrachiatum, T. neokoningii, T. pseudokoningii, and T. yunnanens) are effective biocontrol agents due to their ability to inhibit pathogen growth in soil (Atanasova et al. 2013; Kredics et al. 2014; Parmar et al. 2015). These species help to the overall of the plant antagonistic microorganisms. Trichoderma spp. reduce the growth and survival of pathogens through various mechanisms such as competition antibiosis, mycoparasitism, hyphal interactions, and enzyme secretion. Such microorganisms are now available commercially and are used in crop management and practices (Atanasova et al. 2013; Parmar et al. 2015). There are many difficulties in the morphological identification of Trichoderma due to the homoplasy of morphological and phenetic characters, particularly among the Trichoderma anamorph forms; hence, the molecular methods offer a dependable way to identify Trichoderma species (Hassan et al. 2014; Lakhani et al. 2016). Molecular identification and characterization of fungal genomes are generally performed with a universal set of internal transcribed spacer (ITS) markers that amplify the specified 5.8S rRNA fragment of the fungal DNA. The ITS region of nuclear DNA is currently selected as the standard marker for fungal DNA barcoding and for analyzing fungal diversity (Schoch et al. 2012; Hassan 2014). Hassan et al. (2019) improved the use of sequence-characterized amplified region (SCAR) markers as a simple marker system that enables rapid and inexpensive species identification which contributed to overcome Trichoderma identification difficulties. Molecular identification of fungal species using molecular techniques such as DNA barcoding has gained considerable attention; therefore, the International Union of Microbiological Societies hosts an official website, namely the International Sub-commission on Trichoderma and Hypocrea Taxonomy (http://isth.info/). This website is designed as an open resource to host the most recent data on Hypocrea/Trichoderma taxonomy and evolution. Additionally, this website presents a collection of user-friendly tools for the rapid molecular identification of Hypocrea/Trichoderma based on DNA barcode. The TrichOKEY and TrichoBLAST databases are available on the website, which can assist in the quick identification of Trichoderma/Hypocrea species, using ITS anchors (Srivastava et al. 2014; Fahmi et al. 2016).
To date, a limited number of ITS gene sequence are used for the molecular identification of the fungal genus Trichoderma. However, the use of these gene sequences is limited to the successful and accurate identification of only a few species (Hassan 2014; Fahmi et al. 2016; Hassan et al. 2019). Moreover, few studies dealt with the genetic diversity of Trichoderma in Egypt and Saudi Arabia.
The present study is intended to provide novel combinations of ITS primers that are both species- and strain-specific. Also, ITS1 and ITS4 region sequences will be utilized to identifyTrichoderma species, assess its genetic diversity, and determine if the geographic locations have a probable role in shaping Trichoderma’s genetic diversity.
Materials and methods
A total of 36 Trichoderma isolates were collected from Egypt and Saudi Arabia across various geographic locations (Fig. 1). Potato dextrose agar (PDA) medium was used to culture the Trichoderma isolates, and it is containing 4 g potato extract, 20 g dextrose, and 15 g agar.
DNA extraction, polymerase chain reaction, and electrophoresis
Genomic DNA isolation
Trichoderma isolates were cultured on Czapek Dox broth at 28 °C for 5 days. Czapek Dox broth media contain 30 g sucrose, 3 g sodium nitrate, 1 g dipotassium phosphate, 0.5 g magnesium sulfate, 0.5 g potassium chloride, and 0.01 g ferrous sulfate, and pH was adjusted to 7.3. The total genomic DNA for each strain was extracted, using the Norgen Plant/Fungi DNA Isolation Kit (Sigma, Thorold, Canada) as previously described in Hassan et al. (2019). The genomic DNA was subsequently used for sequencing.
PCR amplification of ITS region
ITS1 and ITS4 regions at the 5.8S rDNA gene from all specimens were amplified, using the primer pair ITS-1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS-4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) as suggested by Hermosa et al. (2000) and Hassan (2014). PCR was performed as described in Hassan et al. (2019), in PCR tubes containing 1 μl (10 ng) of genomic DNA, 12.5 μl of GoTaq® Green Master Mix (Promega, Madison, WI, USA), 1 μl of each primer (20 pmol), and deionized distilled water (up to a total volume of 25 μl). For DNA amplification, the tube contents were heated at 94 °C for 10 min in a C1000 Touch™ Thermal Cycler (Bio-Rad, Munich, Germany). Subsequently, Taq polymerase was added, followed by 35 cycles of 1 min at 94 °C, 45 s at 60 °C, and 1 min at 72 °C, with a final 7-min extension at 72 °C.
DNA sequence analysis
The nucleotide sequences of the 5.8S-ITS region were determined, using the Gene analyzer 3121 sequencing service (Macrogen Co., Seoul, South Korea). The deduced 36 sequences were subjected to the National Center for Biotechnology Information (NCBI) BLAST search tool (http://blast.ncbi.nlm.nih.gov) to detect sequence similarity. Each sequence was individually BLAST searched, where BLAST results with the lowest expected value (e value) which indicates the number of non-chanced alignments were selected. To ensure that BLAST outputs were governed by e values, BLAST algorithm parameters were decreased such that the expected threshold was set to a more stringent value of 1e−6. Alignment and editing of the 36 sequences were carried out using Clustal x version 2 (Larkin et al. 2007).
Phylogenetic analyses
Exploratory data and phylogenetic analyses were carried out using R Project for Statistical Computing (R Core Team 2017). Exploratory data analysis was performed using Seqinr (Charif and Lobry 2007) R package. Phylogenetic analyses were carried out by ape package (Paradis et al. 2004). Reconstruction of the phylogenetic tree was performed using the neighbor-joining method (Nei and Gojobori, 1986).
Nucleotide diversity
To determine the level of sequence variability, the following common genetic parameters were estimated for sequences from specimens originating from Egypt and Saudi Arabia: haplotype diversity (Hd), number of polymorphic sites (S), nucleotide diversity per site (θw), and nucleotide diversity (πT). The DnaSP v5.10 (Librado and Rozas 2009) was used for the above computation.
Isolation-by-distance
The degree of isolation-by-distance (IBD) (Rousset 1997) was assessed by testing the association between geographic and genetic distances for all pairs of isolates within each group of sequences. Pairwise geographical distances among isolates were calculated using the GPS coordinates at the sampling localities in the Geographic Distance Matrix Generator v. 1.2.3 (Ersts 2010). Geographic isolation-by-distance referred to the idea that isolates may be spatially distributed across regions. This term sought to determine if there is a statistically significant relationship between genetic distance and geographic distance and the strength of this relationship. Following the regression method of Rousset (1997), geographic distances were log-transformed and genetic distances were expressed as ΦST. The statistical significance of the associations was assessed based on a Mantel test (Mantel 1967) with 1000 permutations, using GenAlEx 6.5 (Peakall and Smouse 2012).
Results and discussion
Trichoderma species identification
The growth pattern of all 36 Trichoderma strains (24 and 12 isolates from Egypt and Saudi Arabia, respectively) cultured on PDA was monitored, and their proliferation pattern revealed significant changes (data not shown). Oligonucleotide barcode is a powerful tool for the identification of Trichoderma species and is recommended as an alternative or alongside morphological methods (Hassan et al. 2019). Molecular data from ITS sequences provided accurate characterization and identification of isolates. However, hereafter, isolates will be referred to as Saudi Arabian isolates or Egyptian isolates, regardless of the sampling location. Tables 1 and 2 showed the results obtained from NCBI BLAST queries for the 36 isolates.
The criteria used to query sequences aimed to reduce the search space (database). For the 36 isolates queries, E values were 0, which indicated non-chanced alignments of the queries. For Egyptian isolates, the percentage identity with similar species ranged between 84 and 100%, while Saudi Arabian isolates showed similarities with similar species ranged between 97 and 99%. Trichoderma isolates were deposited in a gene bank with accession numbers MK680258–MK680293. Moreover, the NCBI BLAST query of Egyptian isolates resulted in 7 Trichoderma species, namely T. bissettii, T. crassum, T. lentiforme, T. longibrachiatum, T. neokoningii, T. pseudokoningii, and T. yunnanens. Of the 24 Egyptian isolates, 11 were identified as T. lentiforme, 7 were T. longibrachiatum, 2 were T. neokoningii, and only one isolate of each species (T. bissettii, T. crassum, T. pseudokoningi, and T. yunnanense) (Table 1). For the Saudi Arabian isolates, only 4 Trichoderma species were identified, T. longibrachiatum, T. lentiforme, T. simmonsii, and T. pseudokoningii. Of the 12 Saudi Arabian isolates, 8 belonged to T. longibrachiatum, 2 belonged to T. simmonsii, and only one belonged to each of T. pseudokoningii and T. simmonsii (Table 2). Comparing species found among Egyptian isolates with their counterparts in Saudi Arabia, T. simmonsii was found only among Saudi Arabian isolates, and T. bissettii, T. crassum, T. neokoningii, and T. yunnanens only appeared in Egyptian isolates. BLAST search at NCBI was used to match the nucleotide sequences of all eight Trichoderma species for the post-molecular identification validation. This confirmed the selected sequences as specific strains of Trichoderma species (Srivastava et al. 2014; Mazrou et al. 2020). A set of 5 oligonucleotide sequences present in all known Hypocrea/Trichoderma ITS1 - 5.8S RNA - ITS4 sequences, was used in combination to identify the species at a genetic level (Hassan 2014; Hassan et al. 2019). Using molecular markers, particularly those based on DNA sequenced data, was an essential technique to decipher the genetic variability of any species (Nagy et al. 2004; Bellemain et al. 2010). In addition, the interspecies identification of the species from different isolates was carried out in this study. The results showed that although the rDNA ITS sequence was very conservative, there were variations on sequence and length among different isolates, and there was a genetic differentiation at various levels. Therefore, the ITS sequence analysis clearly differentiated between Egyptian and Saudi Arabian species. Similarly, Kuhls et al. (1997) used sequence analysis to differentiate between T. reesei and T. longibrachiatum. Many other researchers have used ITS sequences to identify Trichoderma spp. (Sharma et al. 2009; Shahid et al. 2013). Consequently, variation among individuals of the same species was noticed (Fahmi et al. 2016; Hassan et al. 2019; Mazrou et al. 2020); therefore, it is of great interest to identify the species at the molecular level.
Sequence variations
Sequence length varied greatly among the isolates. For the Egyptian isolates, the shortest sequence length was attributable to T. bissettii of 430 bp, while T. yunnanens had the longest length of 874 bp. The percentage of GC content was consistent among species, with a similar trend observed for the Saudi Arabian isolates. In Egyptian isolates, the GC content ranged from 55% for T. yunnanens to 58% for T. longibrachiatum. Similarly, for the Saudi Arabian isolates, the GC content ranged from 55% in T. simmonsii to 59% in T. pseudokoningii. The base frequencies of Egyptian and Saudi Arabian isolates were presented in Fig. 2a, b. Only 2 species from the Egyptian isolates showed noticeable variation in nucleotide base proportions. Sequences from T. neokoningii and T. yunnanens revealed that the percent of thymine was higher than other bases, and no other noticeable differences were observed. In an earlier study, Abd-Elsalam et al. (2010) observed a slight variation between T. harzianum and T. longibrachiatum in Saudi Arabian isolates. This indicated a wide range of interspecies variations, which is consistent with the idea of haplotype presence among species (Gupta et al. 2014). Abd-Elhamid et al. (2017) also reported that molecular identification is very important to identify the Trichoderma species, so it is recommended to be used for confirming the morphological approaches in the identification of Trichoderma isolates obtained from Egyptian soils.
Phylogenetic analysis
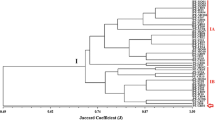
Cluster analysis was carried out as a pre-processing step to glean an insight into the data distribution. The results of the cluster analysis were shown in Fig. 3. The studied 36 isolates formed two large clusters. The first comprised all Saudi Trichoderma strains; this cluster was divided into two sub-clusters. One contained all 8 T. longibrachiatum and T. pseudokoningii, whereas the second comprised T. lentiforme and T. simmonsii. The second cluster comprised all the Egyptian Trichoderma species; this cluster incorporates three sub-clusters. The first comprised T. yunnanens and T. neokoningii. Although the isolates from Saudi Arabia and Egypt were grouped into different groups (Hassan et al. 2014; Fahmi et al. 2016), the Egyptian isolates showed more uniform patterns of clustering. T. lentiforme isolates from Egypt were grouped into one cluster; however, T. crassum was grouped with T. lentiforme Egyptian isolates. These results were similar to those obtained by Anees et al. (2010) in Pakistan, Abd-Elsalam et al. (2010) in Saudi Arabia, Hassan et al. (2014) in Egypt, and Lakhani et al. (2016) in India. Furthermore, T. longibrachiatum grouped with T. pseudokoningii in Saudi Arabian isolates. Besides, T. lentiforme and T. crassum grouped; however, they were subdivided into different subgroups. In an earlier study, Cázares-Garcia et al. (2013) and Hassan et al. (2014) observed a little variation between T. lentiforme and T. longibrachiatum. The low genetic differentiation among isolates from different regions is indicative of shred isolates between the two regions, nevertheless maintaining high genetic variability within each group.
Polymorphism and genetic diversity among species
The number of haplotypes identified among KSA isolates was higher than that of Egyptian isolates (11 and 8, respectively) (Table 3).
Also, the Saudi Arabian isolates showed 98% haplotype diversity compared to 76% among Egyptian isolates. However, the Saudi Arabian isolates showed lower nucleotide diversity (πT) than their Egyptian counterparts.
AMOVA showed significant differentiation between Saudi Arabian and Egyptian isolates with the most genetic variance observed between rather than within regions. Also, it showed a significant genetic differentiation between the two groups (ΦST = 0.838, Table 4).
Most of the genetic variation was attributed to the between-group differences (83.77%). The highest genetic differentiation between isolates in the regions sampled was indicative of the presence of unique species in the two regions. This result was also supported by both cluster and phylogenetic analysis (Fig. 3). Additionally, the statistical analysis revealed further support of the presence of unique isolates where most of the genetic variance explained between the region’s differences (Sharma et al. 2009; Shahid et al. 2013; Mazrou et al. 2020).
Isolation-by-distance
The Mantel test showed no correlation between the genetic and geographic distances in Egyptian and Saudi Arabian isolates (r2 = − 0.38, r2 = 0.23, P > 0.05 for Egyptian and Saudi Arabian isolates, respectively) (Fig. 4a, b). The lack of correlation between genetic and geographic distances was indicative of the absence of isolation-by-distance pattern among isolates from the two regions. The observed pattern suggested the presence of high gene flow within each of the studied regions.
DNA sequencing of the 5.8S-ITS region was carried out, using specific primers within ITS1 and ITS4. The ITS region is one of the most reliable loci for the identification of a strain at the species level (Kullnig-Gradinger et al. 2002; Hassan 2014; Hassan et al. 2019). By comparing the sequences of the 5.8S-ITS region to the sequences deposited in GenBank, all the Trichoderma isolates can be identified to the species level with a homology percentage of at least 99%. The TrichOKEY search tool, a program specifically comparing ITS1 and ITS4 sequences to a specific database for Trichoderma generated from only vouchered sequences, was used to assess the reliability of BLAST results. TrichOKEY has resulted in the successful identification of Trichoderma isolates (Anees et al. 2010; Hassan et al. 2014; Lakhani et al. 2016).
Conclusion
The obtained results have confirmed the efficiency and reproducibility of ITS as a powerful tool for the identification and genetic diversity assessment of Trichoderma spp. Egyptian isolates showed a more uniform phylogenetic pattern than Saudi Arabian isolates. Geographical differences have a significant impact on genetic differentiation of the local isolates where the most of genetic differences occurred among rather than within groups.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- ITS:
-
Internal transcribed spacer
- NCBI:
-
National Center for Biotechnology Information
- BLAST:
-
Basic Local Alignment Search Tool
- AMOVA:
-
Analysis of molecular variance
- IBD:
-
Isolation-by-distance
- PCR:
-
Polymerase chain reaction
References
Abd-Elhamid MI, Ismail MI, Alshishtawy HM, Sedik MZ (2017) Identification and molecular characterization of Egyptian Trichoderma isolates. Biosci res 14:1156–1166
Abd-Elsalam KA, Almohimeed I, Moslem MA, Bahkali AH (2010) M13-microsatellite PCR and rDNA sequence markers for identification of Trichoderma (Hypocreaceae) species in Saudi Arabian soil. Gen Molecul Res 9:2016–2024
Anees M, Tronsmo A, Hermann V, Hjeljord LG, Heraud C, Steinberg C (2010) Characterization of field strains of Trichoderma antagonistic against Rhizoctonia solani. Fungal Biol 114:691–701
Atanasova L, Crom SL, Gruber S, Coulpier F, Seidl-Seiboth V, Kubicek C, Druzhinina I (2013) Comparative transcriptomics reveals different strategies of Trichoderma mycoparasitism. BMC Genomics 14:121–128
Bellemain E, Carlsen T, Brochmann C (2010) ITS as an environmental DNA barcode for fungi: an in-silico approach reveals potential PCR biases. BMC Microbiol 10:189
Cázares-Garcia SV, Vázquez-Garciduenãs MS, Vázquez-Marrufo G (2013) Structural and phylogenetic analysis of laccases from Trichoderma: a bioinformatic approach. PLoS ONE 8:e55295
Charif D, Lobry RJ (2007) SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. Biomed Engin 34:207–232
Ersts PJ (2010) Geographic Distance Matrix Generator v. 1.2.3. Available online: http://biodiversityinformatics.amnh.org/open_source/gdmg
Fahmi AI, Eissa RA, El-Halfawi KA, Hamza HA, Helwa MS (2016) Identification of Trichoderma spp. by DNA barcode and screening for cellulolytic activity. J Microb Biochem Technol 8:202–209
Gupta VG, Schmoll M, Herrera-Estrella A, Upadhyay RS, Druzhinina I (2014) Biotechnology and biology of Trichoderma. Elsevier 5:16–332
Hassan MM (2014) Influence of protoplast fusion between two Trichoderma spp. on extracellular enzymes production and antagonistic activity. Biotechnol Biotechnol Equip 28:1014–1023
Hassan MM, Farid MA, Gaber A (2019) Rapid identification of Trichoderma koningiopsis and Trichoderma longibrachiatum using sequence characterized amplified region markers. Egypt J Biol Pest Control 29:13
Hassan MM, Gaber A, El-Hallous EI (2014) Molecular and morphological characterization of Trichoderma harzianum from different Egyptian soils. Wulfenia J 21:80–96
Hermosa MR, Grondona I, Iturriaga EA, Diaz-Minguez JM, Castro C, Monte E (2000) Molecular characterization and identification of biocontrol isolates of Trichoderma spp. Appl Environ Microbiol 66:1890–1898
Kredics L, Hatvani L, Naeimi S, Körmöczi P, Manczinger L, Vágvölgyi C, Druzhinina I (2014) Biodiversity of the genus Hypocrea/Trichoderma in different habitats. In: Gupta VK, Schmoll M, Herrera-Estrella A, Upadhyay RS, Druzhinina I, Tuohy MG (eds) Biotechnology and biology of Trichoderma. Elsevier, London, pp 3–24
Kuhls K, Lieckfeldt E, Samuels GJ (1997) Revision of Trichoderma section Longibrachiatum including related teleomorphs based on an analysis of ribosomal DNA internal transcribed spacer sequences. Mycologia 89:442–460
Kullnig-Gradinger CM, Szakacs G, Kubicek CP (2002) Phylogeny and evolution of the genus Trichoderma: a multigene approach. Mycol Res 106:757–767
Lakhani HN, Vakharia DN, Hassan MM (2016) Fingerprinting and molecular comparison among two parental strains of Trichoderma spp. and their corresponding fusants produced by protoplast fusion. Biotechnol Biotechnol Equip 30:1065–1074
Larkin MA, Blackshields G, Brown NP, Chenna MGPA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinf 25:1451–1452
Mantel NA (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Mazrou Y, Makhlouf A, Elseehy M, Awad M, Hassan M (2020) Antagonistic activity and molecular characterization of biological control agent Trichoderma harzianum from Saudi Arabia. Egypt J Biol Pest Control 30:4
Nagy E, Kredics L, Antal Z, Papp T (2004) Molecular diagnosis, epidemiology and taxonomy of emerging medically important filamentous fungi. Rev Med Microbiol 15:153–162
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Paradis E, Claude J, Strimmer J (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290
Parmar HJ, Hassan MM, Bodar NP, Umrania VV, Patel SV, Lakhani HN (2015) In vitro antagonism between phytopathogenic fungi Sclerotium rolfsii and Trichoderma strains. Int J Appl Sci Biotechnol 3:16–19
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Schoch CL, Seifert KA, Huhndorf S (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci 109:6241–6246
Shahid M, Srivastava M, Sharma A (2013) Morphological, molecular identification and SSR marker analysis of a potential strain of Trichoderma/Hypocrea for production of a bioformulation. J Plant Pathol Microbiol 4:204–210
Sharma K, Mishra AK, Misra RS (2009) Morphological, biochemical and molecular characterization of Trichoderma harzianum isolates for their efficacy as biocontrol agents. J Phytopathol 157:51–56
Srivastava M, Shahid M, Pandey S, Singh A, Kumar V, Gupta S, Maurya M (2014) Trichoderma genome to genomics: a review. J Data Mining Genomics Proteomics 5:162
Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through the Program of Research Groups under grant number (project number 2/28/1440). The current work was also funded by Taif University Researchers Supporting Project number (TURSP - 2020/59), Taif university, Taif, Saudi Arabia.
Funding
The current work was funded by both King Khalid University ,under grant number (2/28/1440) and by Taif University Researchers Supporting Project number (TURSP -2020/59), Taif university, Taif, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
All authors contributed significantly to this research and preparation of the manuscript. This study was conceived and designed by YM, AM, and MI. The laboratory work was carried out by MH, AB, and AH. Data were analyzed by YM, AS, and MI. The manuscript was written by AK, MI, and AS. The manuscript was revised by AB and AH. All authors have reviewed and agreed upon the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mazrou, Y.S.A., Baazeem, A., Makhlouf, A.H. et al. Comparative molecular genetic diversity between Trichoderma spp. from Egypt and Saudi Arabia. Egypt J Biol Pest Control 30, 120 (2020). https://doi.org/10.1186/s41938-020-00318-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-020-00318-w