Abstract
Background
DNA profiling for the victim identification in several forensic situations is mostly challenging due to a high degree of degradation or due to a high level of intermingling of remains. There are many intrinsic and extrinsic factors involved in the postmortem decay of tissues. Forensic DNA analysts come across a variety of samples ranging from stain samples to visceral material to bone or teeth. The amount and quality of DNA recovery are highly variable/challenging in dealing such forensic evidence.
Material and methods
In the present study, two different types of postmortem tissues (brain and prostate/uterus) along with teeth of 20 naturally decomposed human bodies were analysed. DNA extraction has been performed using the phenol-chloroform extraction method. Further, the extracted samples have been subjected for quantitative analysis using RT-PCR. Samples were amplified using the PowerPlex® 21(PP 21) multiplex system and were genotyped using capillary electrophoresis in 3500XL Genetic Analyzer.
Results
A perfect short tandem repeat (STR) profile has been generated from all the teeth samples. In most of the cases, brain tissue samples also provided an optimal profile. In contrast, prostate tissue samples gave partial amplification at almost all the samples. These findings were further evaluated by DNA profile quality measures for the studied samples.
Conclusion
The present study may provide a platform to effectively evaluate forensic samples that contain small or degraded DNA template. The multiplex autosomal STR loci PP 21 system used in the study is efficient enough for typing such degraded samples with high sensitivity and discrimination power.
Similar content being viewed by others
Background
Personal identification or establishing the identity of an individual is always a challenge for social and medico-legal purposes, especially in cases of natural mass disasters like floods, earthquakes, cases of fire deaths (Heinrich et al. 2009; Dumache et al. 2016), tsunamis, landslides and serious vehicular/train accidents; plane crashes (Hsu et al. 1999); and skeletal remains from mass graves (Prado et al. 1997; Johnston and Stephenson, 2016) or man-made disasters such as terrorist attacks; bomb blasts (Holland et al. 2003); and mass murders also lead to multiple fatalities. In such situations, it is difficult to establish an identity with high accuracy of individuals many a times. Various conventional forensic techniques like fingerprint analysis, anthropology, odontology, etc. have been in use for the identification of human remains. However, there are cases in which the recovered remains are not recognisable by using these conventional methods. Examinations on these material and cases often provide inconsistent and insufficient results due to a high level of commingling of skeletal remains or perhaps due to insufficient ante-mortem data (Gunby 1994). In these circumstances, forensic investigation of biological samples often requires the analysis of human DNA (Alonso et al. 2001). In these kinds of catastrophic events, DNA is expected to undergo a great degree of fragmentation due to natural process resulting from the exposure of the stain or tissue samples to the environment. The level of degradation may get influenced by light, humidity or elevated temperature. The growth of bacterial and fungal contaminations can also lead to physical, chemical and biochemical degradation of genomic DNA (Whitaker et al. 1995; Fondevila et al. 2008). There have been noteworthy advancements in DNA technology used to genotype such fragmented samples especially low DNA amplification and inhibitor tolerance.
The forensically important autosomal short tandem repeat (STR) loci are composed of tandemly repetitive sequences of DNA 2–6 base pairs in length. The regions containing the repeats are generally quite small and therefore undergo possible amplification by PCR (Lygo et al. 1994; Kimpton et al. 1994; Kimpton et al. 1996; Whitaker et al. 1995). Capillary electrophoresis-based DNA profiling using PCR with STR loci is the powerful and widely accepted method for human identification (Edwards et al. 1991, Budowle et al. 2001, Shrivastava et al. 2016).
Postmortem decay of human bodies is considered to be a complex process. It begins with autolysis and putrefaction which further may be followed by aerobic and bacterial decomposition. Stability of DNA in the brain cortex, lymph nodes and psoas muscle can be observed over a period of 3 weeks postmortem (Bar et al. 1988). While teeth and bones serve as a good source of DNA even after a period of months and years (Kaneko et al. 2015). Furthermore, DNA is well preserved in teeth as compared to other bones (Woodward et al., 1994). The advancement of decomposition of a corpse depends on a number of intrinsic and extrinsic factors (Adlam and Simmons, 2007; Duday and Guillon, 2006; Hau et al. 2014). It is generally considered that head and limbs decompose at a faster rate as compared to the body trunk, mainly due to the presence of a higher mass of soft tissue (Braig and Perotti, 2009; Matuszewski et al. 2008). The prostate or uterus are considered to last long or resists decomposition for longer duration (Pinheiro, 2006). However, the rate of degradation varies from body to body and also depends on a number of environmental factors. There have been reports regarding the successful PCR-based identification of artificially degraded samples, blood stains or other biological stains (Moretti et al. 2000; Dixon et al. 2006). Keeping in view these points, two different visceral tissues, one prostate tissue, reported to resist degradation for a long time (Pinheiro, 2006), and the other a soft tissue (brain) but reported to retain DNA for a long time (Bar et al. 1988) along with teeth sample, the best source of DNA for identification as reference, are taken into consideration. The present study aims to assess the suitability of two naturally degraded soft tissues and teeth as a reference biological sample for DNA analysis using a new generation autosomal STR multiplex system (PowerPlex 21TM, Promega Corporation).
Materials and methods
Sample collection
Two different types of tissue samples (brain cortex and prostate/uterus tissue) and teeth samples were collected from 20 unclaimed decomposed bodies with post-mortem interval up to 25 days submitted at the mortuary of All India Institute of Medical Sciences (AIIMS), New Delhi. The tissue samples were collected and stored in sterilised plastic containers at − 80 °C until further processing. No preservative was used to store the samples. Majority of the brain tissue samples were in a liquefied state at the time of collection itself while the prostate tissues were all intact. Teeth samples were buried in garden soil for a minimum period of 10 months prior to extraction.
DNA extraction and quantification
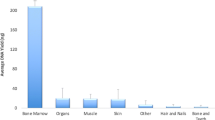
The DNA was extracted using the phenol-chloroform procedure (Sambrook et al. 1989) with required modifications. All the samples were eluted in 15 μl of TE buffer and stored at − 20 °C till further analysis. Each DNA extract was quantified by a real-time PCR using Quantifiler® Trio DNA Quantification kit (Thermo Fisher Scientific, Oyster Point, CA) (lot no. 1607013), with an ABI Prism 7500 detection system (Applied Biosysytems) according to manufacturer’s recommendations (https://assets.thermofisher.com/TFS-Assets/LSG/manuals/4485354.pdf), except half-reaction volume was used (Cho et al. 2018). Internal positive controls (IPCs) were used to monitor PCR inhibitors during the reaction. Also, negative controls and reagent blanks were included in every step of the study.
PCR amplification
PCR amplification was performed on the Gene Amp PCR system 9700 (Applied Biosystems). The extracted DNA samples were amplified with Promega PowerPlex 21TM STR kit (lot no. 0000309091), which simultaneously amplifies 20 autosomal STR loci (D3S1358, D1S1656, D6S1043, D13S317, Penta E, D16S539, D18S51, D2S1338, CSF1PO, Penta D, TH01, vWA, D21S11, D7S820, D5S818, TPOX, D8S1179, D12S391, D19S433, FGA) and amelogenin marker. Amplification was done as per the manufacturer’s protocol except half-reaction volume was used (Alenizi et al. 2009; Batham et al. 2019). A total of 8 μl of final reaction mix was used for PCR with 0.5 μl of extracted DNA.
DNA detection and data analyses
Capillary electrophoresis was performed using the 3500XL Genetic Analyzer System (Applied Biosystems). A total of 10 μl of sample injection volume was used with 1 μl of PCR product, 10 μl of the Hi-Di formamide (lot no. 1710526) and 0.2 μl of the WEN ILS 500 Size standard (lot no. 0000309091). Detection of amplified STRs was carried out on ABI Prism 3500XL genetic analyzer. Data analysis was carried out using GeneMapper ID v3.2 software. Alleles were designated on the basis of the number of allele repeats with the help of an allelic ladder (lot no. 0000309091) provided by the manufacturers of the multiplex systems used for the study. Peak detection threshold was set to 50 RFUs for allelic designation.
Results
The results showed that the success rate of brain samples is found better than prostate samples. Brain tissue (Fig. 1) and teeth sample (Fig. 2) gave full profile at all the 21 STR markers in 16 and 9 samples, respectively, unlike prostate tissue (Fig. 3) which showed partial amplification at a majority of the loci of PowerPlex 21and no prostate sample yielded a complete profile. The genotyping results are also supported by quantification results obtained by using RT-PCR (Additional file 1: Tables S1, S2, and S3).
The total number of amplified loci per sample for each of the three types of tested biological samples in the study is presented in Table 1. An interesting finding with brain tissue is the fact that only two of the tested samples yielded non-amplifiable DNA. In more than 12 samples, 15 or more loci were amplified and allele dropout mostly occurred on the loci which are more than 300 base pairs namely D13S317, Penta D, Penta E, CSF1PO, TPOX and FGA loci. Most of the other amplified loci in all the samples passed DNA profile quality measures. In the case of prostate tissues, not even a single sample was successful in providing a full profile at all 21 loci. Only six of the samples showed amplification at more than 15 loci while the remaining 14 samples showed dropouts in almost 15 loci on an average. In contrast, teeth samples exhibit full profile at all 21 loci in 16 tested samples. Only four samples showed dropout at D2S1338, TPOX and FGA loci which are also the larger base pair loci (265–411 bp). The occurrence of each of the 21 STR markers in brain tissue, prostate tissue and teeth samples has been shown graphically (Fig. 4).
Profile quality
DNA profile quality of the samples which showed amplification at all the 21 markers has been evaluated by estimating the following measures: balance between allelic peaks within heterozygous loci (intra-locus or local balance), total allelic peak height (TPH) and inter-colour balance between STR markers (ICB or global balance) using PRISM statistical software. In the case of brain tissue samples, the mean value for peak height ratio (PHR) ranges from 0.62 to 0.92 which indicates most of the samples have PHR near to the standard value of 0.7. Also, lower 95% confidence interval of mean ranges from 0.39 to 0.86 while upper 95% confidence interval of mean ranges from 0.78 to 1.1 (Fig. 5a). Likewise, the mean value for TPH (total peak height) ranges from 1144 to 27,812. Also, lower 95% confidence interval of mean ranges from 343 to 13,954 while upper 95% confidence interval of mean ranges from 2876 to 42,984 (Fig. 5b). Similarly, the mean value for global balance ranges from 38,800 to 60,281. Also, lower 95% confidence interval of mean ranges from 20,520 to 37,092 while upper 95% confidence interval of mean ranges from 57,080 to 84,709 (Fig. 5c). In the case of teeth samples, the mean value for PHR (peak height ratio) ranges from 0.72 to 0.85 which indicates that all of the samples have PHR near to the standard value of 0.7. Also, lower 95% confidence interval of mean ranges from 0.66 to 0.83 while upper 95% confidence interval of mean ranges from 0.84 to 0.93 (Fig. 6a). Likewise, the mean value for TPH (total peak height) ranges from 1144 to 23,967. Also, lower 95% confidence interval of mean ranges from 343 to 13954 while upper 95% confidence interval of mean ranges from 1649 to 42,984 (Fig. 6b). Similarly, the mean value for global balance ranges from 57,213 to 106,779. Also, lower 95% confidence interval of mean ranges from 42,021 to 83341 while upper 95% confidence interval of mean ranges from 72,404 to 130217 (Fig. 6c). These measures were not evaluated for prostate tissue as none of the samples showed amplification at all the 21 STR loci.
Discussions
We demonstrated the PCR amplification of STR loci from decomposed biological samples. The high success rate for nuclear STR typing reported here further confirmed that STRs could be considered a method of choice in casework involving rarely sampled tissues (brain and prostate). The study also suggests the use of brain tissue as amplifiable DNA was detected in 18 of the total 20 samples tested. The results also show that peak height ratio, inter-colour balance and total peak height (which are profile quality parameters) observed in teeth are in agreement with the brain tissue. No amplification or dropout on the larger loci (more than 250 base pairs) included in the multiplex system suggest the need of the use of specifically designed kits for degraded DNA to achieve the amplification at these larger loci such as ABI Minifiler kit (Lopes et al. 2009; Welch et al. 2011; Constantinescu et al. 2012). In this study, the effects of environmental variables were not controlled, since the samples were collected from a wide range of contexts. In general, the results are consistent with those reported in other studies where DNA profiling was performed using different tissues of decomposed human bodies. These study involved DNA amplification of STR loci from a relative amount of DNA present in the heart, liver, femoral muscles and thigh muscles, respectively (Bar et al. 1988; Clayton et al. 1995; Takahashia et al., 1997).
The results in this study are categorised as per the suitability of samples among the brain, prostate/uterus and teeth for the purpose of individualization. This provides information on the amplification efficiency of different types of decomposed biological samples considered in the study. Although teeth are preferred over soft tissues since its robust nature preserves DNA for a longer duration (Johnston and Stephenson 2016). Even after being buried in the garden soil for the duration of 10 months, no substantial difference could be seen in the profile quality of teeth samples. Since it is evident from the results, attained by using PRISM software that brain tissue samples which showed amplification at all loci may also correspond to a good profile quality as obtained by teeth samples. These profile quality measures further provide definitive verification that brain tissue and teeth samples could be considered as optimal samples for DNA recovery. But in situations where the body is found in a skeletonised form, or in a highly putrefied state, teeth can be considered as the best samples for identification purposes (Alonso et al. 2005). Also, with the continual advancements in capillary electrophoresis-based STR marker multiplex systems, six dye chemistry-based multiplex kits are also available recently. Improved chemistries of different multiplex systems may result into amplification of brain tissues on more markers. Improved methods of extraction including automated DNA extractions may further be tested on these samples.
Conclusion
The present study put forward the use of brain tissue samples, which can provide comparable results with that of teeth samples, when collected and preserved in a proper manner. In contrast, being the last organ to decompose, prostate or uterus tissue could not be a reliable source for DNA profiling. Teeth samples usually take a longer duration of processing time as compared to tissue samples. However, larger environmental interferences are yet to be explored for its suitability in different kinds of forensic situations. The study concludes that human brain tissues can also serve the purpose of identification to a good extent even from the bodies recovered in a decomposed state where it could not get identified just by seeing the facial features. Although, in the cases of skeletonised remains, teeth samples still remain the sample of choice.
Availability of data and materials
The materials/samples are obtained by the unclaimed putrefied human corpse submitted at AIIMS mortuary after obtaining ethical approval for the study.
Abbreviations
- ILS:
-
Internal lane standard
- PCR:
-
Polymerase chain reaction
- RFU:
-
Relative fluorescence units
- RT-PCR:
-
Real-time PCR
References
Adlam RE and Simmons T (2007) The effect of repeated physical disturbance on soft tissue decomposition-are taphonomic studies an accurate reflection of decomposition. J Forensic Sci 52(5):1007–1014
Alenizi MA, Goodwin W, Hadi S, Alenizi HH, Altamar KA, Alsikel MS (2009) Concordance between the AmpF‘STR MiniFilerTM and AmpF‘STR Identifiler PCR amplification kits in the Kuwaiti population. J Forensic Sci 54(2):350–352
Alonso A, Andelinovic S, Martin P, Sutlovic D, Erceg I, Huffine E (2001) DNA typing from skeletal remains: evaluation of multiplex and megaplex STR systems on DNA isolated from bone and teeth samples. Croat Med J 42(3):260–266
Alonso A, Martín P, Albarrán C, García P, Fernández de Simón L, Iturralde MJ, Fernández-Rodríguez A, Atienza I, Capilla J, García-Hirschfeld J, Martínez P, Vallejo G, García O, García E, Real P, Álvarez D, León A, Sancho M (2005) Challenges of DNA profiling in mass disaster investigations. Croat Med J 46(4):540–548
Bar W, Kratzer A, Machler M, Schmid W (1988) Postmortem stability of DNA. Forensic Sci Int 39:59–70
Batham MS, Kushwaha KPS, Chauhan T, Kumawat RK, Shrivastava P (2019) Autosomal STR allele frequencies in Kahars of Uttar Pradesh, India, drawn with PowerPlex® 21 multiplex system. Int J Leg Med. https://doi.org/10.1007/s00414-019-02046-9
Braig HR and Perotti MA (2009) Carcases and mites. Exp Appl Acarol 49(1):45–84
Budowle B, Shea B, Niezgoda S, Chakraborty R (2001) CODIS STR loci data from 41 sample populations. J Forensic Sci. 46:453–489
Clayton TM, Whitaker JP, Fisherb DL, Leeb DA, Holland MM, Weednb VW, Maguire CN, DiZinno JA, Kimptond CP and Gill P. (1995): Further validation of a quadruplex STR DNA typing system: a collaborative effort to identify victims of a mass disaster. Forensic Sci. Int. 76:17–25.
Cho Y, Kim HS, Kim MH, Park M, Kwon H, Lee YH, Lee DS (2018) Validation of reduced reagent volumes in the implementation of the Quantifiler Trio Quantification kit. J Forensic Sci 63(2):517–525
Constantinescu CM, Barbarii LE, Iancu CB, Constantinescu A, Neagu E, Iancu D, Girbea G (2012) Challenging DNA samples solved with MiniSTR analysis. Rom J Leg Med 20:51–56
Dixon LA, Dobbins AE, Pulker HK, Butler JM, Vallone PM, Coble MD, Parson W, Berger B, Grubwieser P, Mogensen HS, Morling N, Nielsen K, Sanchez JJ, Petkovski E, Carracedo A, Sanchez-Diz P, Ramos-Luis E, Brio M, Irwin JA, Just RS, Loreille O, Parsons TJ, Syndercombe-Court D, Schmitter H, Stradmann-Bellinghausen B, Bender K, Gill P (2006) Analysis of artificially degraded DNA using STRs and SNPs-results of a collaborative European (EDNAP) exercise. Forensic Sci. Int. 164:33–44
Duday H, Guillon M (2006) Understanding the circumstances of decomposition when the body is skeletonized. In: Schmitt A, Cunha E, Pinheiro J (eds) Forensic anthropology and medicine: complimentary sciences from recovery to cause of death. Humana Press Inc, Totowa, pp 117–157
Dumache R, Muresan C, Ciocan V, Rogobete AF, Enache A (2016) Post-mortem identification of a fire carbonized body by STR genotyping. Clin Lab 62(10):2033–2037
Edwards A, Civitello A, Hammond HA, Caskey CT (1991) DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am J Hum Genet 49:746–756
Fondevila M, Phillips C, Naveran N, Cerezo M, Rodriguez A, Calvo R, Fernandez LM, Carracedo A, Lareu MV (2008) Challenging DNA: assessment of a range of genotyping approaches for highly degraded forensic samples. FSI: Genet Supp Ser 1:26–28
Gunby P (1994) Medical team seeks to identify human remains from mass graves of war in former Yugoslavia. JAMA 272:1804–1806
Hau TC, Hamzah NH, Lian HH, Hamzah SPAA (2014) Decomposition process and post mortem changes: review. Sains Malaysiana 43(12):1873–1882
Heinrich A, Schwark T, Simeoni E, von Wurmb-Schwark N (2009) Genetic identification of fire deaths. Forensic Sci Int Genet 2:s253–s254
Holland MM, Cave CA, Holland CA, Bille TW (2003) Development of a quality, high throughput DNA analysis procedure for skeletal samples to assist with the identification of victims from the World Trade Centre attacks. Croat Med J 44(3):24–272
Hsu CM, Huang NE, Tsai LC, Kao LG, Chao CH, Linacre A, Lee JCI (1999) Identification of victims of the 1988 Taoyuan Airbus crash accident using DNA analysis. Int J Legal Med 113:43–46
Johnston E and Stephenson M (2016) DNA profiling success rates from degraded skeletal remains in Guatemala. J Forensic Sci 61(4):898–902
Kaneko Y, Ohira H, Tsuda Y, Yamada Y (2015) Comparison of hard tissues that are useful for DNA analysis in forensic autopsy. Leg Med 17(6):547–552
Kimpton CP, Fisher D, Watson S, Adams M, Urquhart A, Lygo J, Gill P (1994) Evaluation of an automated DNA profiling system employing multiplex amplification of four tetrameric STR loci. Int J Legal Med 106:302–311
Kimpton CP, Oldroyd NJ, Watson SK, Frazier RER, Johnson PE, Milican ES, Urquhart A, Sparkes RL, Gill PD (1996) Validation of a highly discriminating multiplex short tandem repeat amplification system for human identification. Electrophoresis 17:1283–1293
Lopes V, Andrade L, Carvalho M, Serra A, Balsa F, Bento AM, Batista L, Oliveira C, Corte-Real F, Anjos MJ (2009) Mini-STRs: A powerful tool to identify genetic profiles in samples with small amounts of DNA. FSI: Genet Supp Ser 2:121–122
Lygo JE, Johnson PE, Holdaway DJ, Woodroffe S, Whitaker JP, Clayton TM, Kimpton CP, Gill P (1994) The validation of short tandem repeat (STR) loci for use in forensic casework. Int J Leg Med 107:77–89
Matuszewski S, Bajerlein D, Konwerski S, Szpila K (2008) An initial study of insect succession and carrion decomposition in various forest habitats of central Europe. Forensic Sci Int 180(23):61–69
Moretti TR, Baumstark AL, Defenbaugh DA, Keys KM, Smerick JB, Budowle B (2000) Validation of short tandem repeats (STRs) for forensic usage: performance testing of fluorescent multiplex STR systems and analysis of authentic and simulated forensic samples. J Forensic Sci 46(3):647–660
Pinheiro J (2006) Decay Process of a Cadaver. In: Schmitt A, Cunha E, Pinheiro J (eds) Forensic anthropology and medicine. Humana Press Inc, Totowa
Prado VF, Castro AK, Oliveira CL, Souza KT, Pena SD (1997) Extraction of DNA from human skeletal remains: practical applications in forensic sciences. Genet Anal 14:41–44
Sambrook J, Fritsch EF and Maniatis T (1989) Molecular Cloning: A Laboratory Manual. 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Shrivastava P, Jain T, Trivedi VB (2016) DNA fingerprinting: a substantial and imperative aid to forensic investigation. Eur J Forensic Sci 3(3):23–30
Takahashia M, Katoa Y, Mukoyamab H, Kanaya H, Kamiyama S (1997) Evaluation of five polymorphic microsatellite markers for typing DNA from decomposed human tissues–correlation between the size of the alleles and that of the template DNA. Forensic Sci. Int. 90:1–9
Welch L, Gill P, Schneider PM, Parson W, Mogensen HS, Morling N (2011) A comparison of mini-STRs versus standard STRs-results of a collaborative European (EDNAP) exercise. Forensic Sci Int Genet 5:257–258
Whitaker JP, Clayton TM, Urquhart AJ, Millican ES, Downes TJ, Kimpton CP, Gill P (1995) Short tandem repeat typing of bodies from a mass disaster: high success rate and characteristic amplification patterns in highly degraded samples. BioTechniques 18:670–677
Woodward SR, King MJ, Chiu NM, Kuchar MJ, Griggs CW (1994) Amplification of ancient nuclear DNA from teeth and soft tissues. PCR methods Appl 3:244–247
Acknowledgements
The authors are thankful to Director, State Forensic Science Laboratory, Sagar, MP, India, for providing laboratory facility and Promega (India), for providing the PowerPlex 21TM STR multiplex kit for this study.
Funding
The study was supported by the UGC-JRF fellowship sanctioned to Bhawana Joshi vide letter no F.15-6(Dec.2013)/2014 (NET) dated 02-07-2014.
Author information
Authors and Affiliations
Contributions
MKT, BJ and AR contributed to the study conception and design. SL, AR and BJ contributed to the acquisition of samples. MKT, BJ, PS and AR contributed to the analysis and interpretation of the data. MKT, BJ, AR and PS contributed to the drafting of the manuscript. MKT, BJ, AR and PS contributed to the critical revision of the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval has been obtained from the ethical committee of AIIMS, New Delhi, via letter no. IEC/NP-286/10.07.2015, RP-40/2015, and the ethical committee of Punjabi University, Patiala, via letter no. IEC/26/21.01.16.
Consent for publication
Given by all the authors
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1 Quantification data of brain samples using RT-PCR. Table S2 Quantification data of teeth samples using RT-PCR. Table S3 Quantification data of prostate/uterus samples using RT-PCR. (DOCX 22 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Thakar, M.K., Joshi, B., Shrivastava, P. et al. An assessment of preserved DNA in decomposed biological materials by using forensic DNA profiling. Egypt J Forensic Sci 9, 46 (2019). https://doi.org/10.1186/s41935-019-0151-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41935-019-0151-5