Abstract
Background
To evaluate the 514–524 (CA)n dinucleotide repeats/allele of the mtDNA hypervariable region III in the Urali Kuruman tribe of Southern India. Blood samples were randomly collected from 100 healthy unrelated individuals, and complete mtDNA control region was amplified and sequenced by the Sanger sequencing method.
Findings
The present study revealed the presence of nucleotide insertion and deletion of C and A (CA/AC) at position 514–524 in 86 samples. Insertion of 1AC (14%) and 2AC (20%) and deletion of 1CA (49%) and 2CA (3%) were detected. A statistical estimate for this population showed a genetic diversity of 0.6866 and random match probability of 0.3202, and the discrimination power calculated was 0.6798.
Conclusion
This study will be beneficial for personal identification and in forensic investigation casework.
Similar content being viewed by others
Background
The non-coding or control region, synonymously referred as the displacement loop (D-loop) segment of mitochondrial DNA (mtDNA) is most extensively studied in forensics, particularly the hypervariable regions (HVR I, II, and III) (Wilson et al. 1995; Szibor et al. 1997; Lutz et al. 2000; Chung et al. 2005; Lander et al. 2008; Irwin et al. 2009; Elmadawy et al. 2013; Hayat et al. 2015; Chaitanya et al. 2016; Bhatti et al. 2018). The hypervariable regions are mutation hotspots, which evolve ~ 10 times rapidly than chromosomal DNA, with relatively higher mutation rate giving rise to more polymorphic sites (Stoneking 2000). High copy number and circular form of mtDNA makes it less vulnerable to exonuclease degradation, which makes it a beneficial tool in identifying persons from the highly degraded biological sample in forensic cases (Parson et al. 1998; Thongngam et al. 2016).
Although the HVR I (np 16,024 to 16,365) and HVR II (np 00073 to 00340) regions are unanimously used in forensics, other regions of the D-loop such as the HVR III region (np 00438 to 00574), which was first identified by Lutz and co-workers (Lutz et al. 1997) can also be analyzed as an additional marker which increases the discrimination power of common haplotypes and helps confirm relationships between individuals presenting only one difference in the sequences (Szibor et al. 2007; Nilsson et al. 2008). In addition to point mutations, repetition of nucleotides :single and dinucleotide repeatsoccurs in the D-loop segment known as length heteroplasmy (Li et al. 2016). Commonly occurring length heteroplasmies are the Poly-C (Poly-cytosine) residues at np 16,189 in HVR I, np 303–315 in HVR II, and np 568–573 in HVR III and the five pairs of CA/AC residues at np 514–524 in the HVR III region (Bhatti et al. 2018).
The mitochondrial CA/AC residues are synonymously referred as (CA)n repeats found within the HVR III region are length heteroplasmy or variables similar to the chromosomal DNA short tandem repeats (STRs); mutations at this position occur as an insertion or deletion (indels) of one or more pairs of CA/AC residues. Chromosomal STRs are profoundly used as a forensic marker, and when relating to the sensitivity of D-loop, the (CA)n repeat amplification seems to be a useful technique than the chromosomal STRs of similar fragment length, due to its high copy number and high stability of exonuclease degradation of corpses; the mtDNA D-loop (HVR I, II, and III) analysis is often helpful when chromosomal STRs fail (Szibor et al. 1997); this has already been proven by the examination of mtDNA in skeletal and ancient human samples (Holland et al. 1993; Kurosaki et al. 1993). Therefore, the length heteroplasmy acts as an additional marker in forensic analysis and is very informative in the interpretation of matches between subjects and their maternal relatives (Naue et al. 2015) and can deliver further insights of discriminative potential that helps to identify human remains during forensic casework (Santos et al. 2008).
Nevertheless, the HVR III has proven to be useful as a supplementary polymorphic segment for the forensic application (Hoong and Lek 2005). Even though the majority of forensic cases utilize only the HVR I and HVR II sequences, it is noteworthy that HVR III (CA)n repeat segment has equal potential as an additional mtDNA marker in forensic investigations (Ivanov et al. 1996; Lutz et al. 2000; Chung et al. 2005; Fridman and Gonzalez 2009; Bhatti et al. 2018). No such study of (CA)n dinucleotide indels of mtDNA HVR III 514–524 region in Indian population has been reported so far; this study was aimed to evaluate the (CA)n dinucleotide repeats in the Urali Kuruman tribe of Kerala, South India.
Tribe
Urali Kurumans are an endogamous tribal group who migrated along the unpastoral lands inhabiting the hillocks of Wayanad, Kerala, South India, with a population of approximately 11,179 people (Chandramouli- Census of India 2011). Urali Kurumans are slender to medium in stature and dark brown or dark in complexion, and some members of the tribe exhibit platyrrhine features; they are monogamous and neolocal and speak a dialect of the Dravidian language family. Traditionally, they were nomadic hunter–gatherers living on steep edges, practicing shift cultivation, foraging, and trapping small birds and animals for food and hide (Singh 2002). Until now, none of the studies have described the genetic structure of the Urali Kuruman tribe. Even though the present study only focusses on the (CA)n dinucleotide repeat of mtDNA HVR III 514–524 region, which will be an impending aspect for studying complete mtDNA and autosomal DNA of the Urali Kurumans.
Materials and methods
Sample collection
Prior to collecting blood samples, written informed consents were obtained from all the 100 healthy unrelated individuals of the Urali Kuruman tribal population of Wayanad, Kerala. About 5–10 ml of sample blood was drawn in vacutainers as per standard protocols.
DNA extraction and quantitation
DNA was extracted following Proteinase-K digestion followed by phenol–chloroform extraction and ethanol precipitation. The extracted DNA was quantified through spectrophotometer (Hitachi U-1800) at an absorption of 260 nm and 280 nm followed by 1% agarose gel electrophoresis (Sambrooke et al. 1989).
Mitochondrial DNA control region analysis
Complete mtDNA control region was amplified using a set of primers illustrated in Table 1. PCR amplification was carried out in a 10μl reaction mixture, having 1 μl of genomic DNA, dNTP 0.5 μl, PCR buffer with MgCl2 1 μl, 0.1 μl of Taq polymerase; 0.2 μl of primers (both forward and reverse); and 7.04 μl of Milli Q using Gene Amp PCR system 9700 thermal cycler (Applied Biosystems) with cycling conditions of 94 °C for 2 min, 95 °C for 1 min, 58 °C for 0.45 s, 72 °C for 2.30 min, and final extension at 72 °C for 7.00 min for 35 cycles. PCR amplicons were cycle sequenced with Big Dye cycle sequencing ready reaction kit (Applied Biosystems), and fluorescent amplimers were detected through ABI 3730 DNA analyzer (Applied Biosystems).
DNA sequence and statistical analysis
Complete control region sequences were analyzed using Seqscape v2.5 (Applied Biosystems) and mutations were scored using Revised Cambridge Reference Sequence (rCRS) (Andrews et al. 1999), and only the (CA)n dinucleotide repeat sequences from position 514–524 of HVR III region are included in this paper. The genetic diversity (h) of the complete (CA)n heteroplasmy was estimated following h = n (1 − ΣX2)/(n − 1), where n is the sample size and X is the frequency of observed haplotypes, and the discrimination power (DP) was also determined using the equation DP = (1 − ΣX2), where X is the frequency of each observed haplotype (Tajima 1989). The random match probability (p) was defined by the equation p = ΣX2, where X is the frequency of each observed haplotype (Stoneking et al. 1991).
Results and discussion
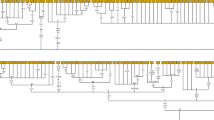
In order to investigate the HVR III (CA)n dinucleotide repeats at position 514–524, the sequences were categorized into nucleotide insertions and deletions, which generally occurs by one or more insertion or deletion of CA/AC nucleotides. The results revealed 86 out of 100 individuals have indel changes in the (CA)5 dinucleotide repeat cluster which is the reference sequence and detected four different (CA)n dinucleotide repeat sequences (Table 2). (CA)5, the threshold sequence, was detected in 14% Urali Kurumans, and (CA)4 repeat was the most frequently detected repeat in 49% Urali Kurumans which occurs by 1 CA deletion. In addition, (CA)6 repeat with 1 AC insertion was detected in 14% Urali Kurumans, and (CA)7 with 2 AC insertion was detected in 20% Urali Kurumans. Likewise, a rare dinucleotide repeat (CA)3 was detected in 3% Urali Kurumans, which occurs when 2 CA nucleotides are deleted, and according to previous reports, (CA)3 was detected in 2% Cameroonians (Szibor et al. 1997) and 1% of Thais (Thongngam et al. 2016). Nevertheless, (CA)4 repeat was also the most frequently detected dinucleotide change in some world populations, and (CA)6 and (CA)7 were also detected in many populations (Table 3). A statistical estimate showed a genetic diversity (h) of 0.6866, a probability of the random match (p) of two individuals sharing the same haplotype was 0.3202, and discrimination power (DP) calculated was 0.6798 in the Urali Kuruman population. (CA)n dinucleotide repeat polymorphisms present in this study and some world populations are inferred in (Table 3), and the sequence structure of the (CA)n dinucleotide detected in this study are depicted in (Fig. 1).
The five (CA)n dinucleotide repeats: a (CA)3 dinucleotide repeat, b (CA)4 dinucleotide repeat, c (CA)5 dinucleotide repeat(rCRS), d (CA)6 dinucleotide repeat, and e (CA)7 dinucleotide repeat genotyped by deletion and insertion of CA/AC nucleotides at position 514–524 of mtDNA HVR III region by Sanger sequencing. Electropherograms of (CA)n dinucleotide repeats at position 514–524 of mtDNA HVR III region detected in the present study
Conclusion
The findings demonstrated the degree of insertion–deletion variability of mtDNA HVR III (CA)n dinucleotide repeat sequences at np 514–524 in 100 individuals of the Urali Kuruman tribe. Furthermore, the study imparts the use of HVR III CA indels in human identification, which has an equal potential as HVR I and HVR II regions, and will contribute as an additional forensic investigation marker.
Abbreviations
- AC:
-
Adenine, Cytosine
- CA:
-
Cytosine, Adenine
- D-loop:
-
Displacement loop
- dNTP:
-
Deoxyribonucleotide triphosphate
- HVR:
-
Hypervariable region
- mtDNA:
-
Mitochondrial DNA
- PCR:
-
Polymerase chain reaction
- rCRS:
-
Revised Cambridge Reference Sequence
References
Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147
Bhatti S, Aslam Khan M, Abbas S, Attimonelli M, Gonzalez GR, Aydin HH, de Souza EM (2018) Problems in Mitochondrial DNA forensics: while interpreting length heteroplasmy conundrum of various Sindhi and Baluchi ethnic groups of Pakistan. Mitochondrial DNA Part A 29(4):501–510
Chaitanya L, van Oven M, Brauer S, Zimmermann B, Huber G, Xavier C, Parson W, de Knijff P, Kayser M (2016) High-quality mtDNA control region sequences from 680 individuals sampled across the Netherlands to establish a national forensic mtDNA reference database. Forensic Sci Int Genet 21:158–167
Chandramouli C (2011) Census of India 2011 – a story of innovations. Press Information Bureau, Government of India
Chung U, Lee HY, Yoo JE, Park MJ, Shin KJ (2005) Mitochondrial DNA CA dinucleotide repeats in Koreans: the presence of length heteroplasmy. Int J Legal Med 119:50–53
Elmadawy MA, Nagai A, Gomaa GM, Hegazy HM, Shaaban FE, Bunai Y (2013) Investigation of mtDNA control region sequences in an Egyptian population sample. Legal Med 15(6):338–341
Fridman C, Gonzalez RS (2009) HVIII discrimination power to distinguish HVI and HVII common sequences. Forensic Sci Int Genet Suppl Ser 2(1):320–321
Haslindawaty AR, Panneerchelvam S, Edinur HA, Norazmi MN, Zafarina Z (2010) Sequence polymorphisms of mtDNA HV1, HV2, and HV3 regions in the Malay population of Peninsular Malaysia. Int J Legal Med 124:415–426
Hayat S, Akhtar T, Siddiqi MH, Rakha A, Haider N, Tayyab M, Abbas G, Ali A, Bokhari SYA, Tariq MA (2015) Mitochondrial DNA control region sequences study in Saraiki population from Pakistan. Leg Med (Tokyo) 17:140–144
Holland MM, Fisher DL, Mitchell LG, Rodriquez WC, Canik JJ, Merril CR, Weedn VW (1993) Mitochondrial DNA sequence analysis of human skeletal remains: identification of remains from the Vietnam War. J Forensic Sci 38(3):542–553
Hoong LL, Lek KC (2005) Genetic polymorphisms in mitochondrial DNA hypervariable regions I, II and III of the Malaysian population. Asia-Pacific J Mol Biol Biotechnol 13:79e85
Irwin JA, Saunier JL, Beh P, Strouss KM, Paintner CD, Parsons TJ (2009) Mitochondrial DNA control region variation in a population sample from Hong Kong, China. Forensic Sci Int Genet 3(4):e119–e125
Ivanov PL, Wadhams MJ, Roby RK, Holland MM, Weedn VW, Parsons TJ (1996) Mitochondrial DNA sequence heteroplasmy in the Grand Duke of Russia Georgij Romanov establishes the authenticity of the remains of Tsar Nicholas II. Nature Genet 12(4):417
Kurosaki K, Matsushita T, Ueda S (1993) Individual DNA identification from ancient human remains. Am J Hum Genet 53(3):638
Lander N, Rojas MG, Chiurillo MA, Ramírez JL (2008) Haplotype diversity in human mitochondrial DNA hypervariable regions I-III in the city of Caracas (Venezuela). Forensic Sci Int Genet 2:e61–ee4
Li M, Rothwell R, Vermaat M, Wachsmuth M, Schröder R, Laros JF, Van Oven M, De Bakker PI, Bovenberg JA, Van Duijn CM, Van Ommen GJ (2016) Transmission of human mtDNA heteroplasmy in the Genome of the Netherlands families: support for a variable-size bottleneck. Genome Res 26:417–426
Lutz S, Weisser HJ, Heizmann J, Pollak S (1997) A third hypervariable region in the human mitochondrial D-loop. Hum Genet 101(3):384
Lutz S, Wittig H, Weisser HJ, Heizmann J, Junge A, Dimo-Simonin N, Parson W, Edelmann J, Anslinger K, Jung S, Augustin C (2000) Is it possible to differentiate mtDNA by means of HVIII in samples that cannot be distinguished by sequencing the HVI and HVII regions? Forensic Sci Int 113(1–3):97–101
Naue J, Horer S, Sanger T, Strobl C, Hatzer-Grubwieser P, Parson W, Lutz-Bonengel S (2015) Evidence for frequent and tissue-specific sequence heteroplasmy in human mitochondrial DNA. Mitochondrion 20:82–94
Nilsson M, Andreasson-Jansson H, Ingman M, Allen M (2008) Evaluation of mitochondrial DNA coding region assays for increased discrimination in forensic analysis. Forensic Sci Int Genet 2(1):1–8
Parson W, Parsons TJ, Scheithauer R, Holland MM (1998) Population data for 101 Austrian Caucasian mitochondrial DNA d-loop sequences: application of mtDNA sequence analysis to a forensic case. Int J Legal Med 111(3):124–132
Rieder MJ, Taylor SL, Tobe VO, Nickerson DA (1998) Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res 26:967–973
Sambrooke J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Santos C, Sierra B, Alvarez L, Ramos A, Fernandez E, Nogués R, Aluja MP (2008) Frequency and pattern of heteroplasmy in the control region of human mitochondrial DNA. J Mol Evol 67(2):191–200
Shneewer HA, Al-loza NG, Kareem MA, Hameed IH (2015) Sequence analysis of mitochondrial DNA hypervariable region III of 400 Iraqi volunteers. Afr J Biotechnol 14(26):2149–2156
Singh KS (2002) People of India: Kerala Vol: XXVII. affiliated east-west press Pvt ltd, New Delhi, pp 749–755
Stoneking M (2000) Hypervariable sites in the mtDNA control region are mutational hotspots. Am J Hum Genet 67(4):1029–1032
Stoneking M, Hedgecock D, Higuchi RG, Vigilant L, Erlich HA (1991) Population variation of human mtDNA control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes. Am J Hum Genet 48:370–382
Szibor R, Michael M, Spitsyn VA, Plate I, Ginter EK, Krause D (1997) Mitochondrial D-loop 3′ (CA)n repeat polymorphism: optimization of analysis and population data. Electrophoresis 18:2857–2860
Szibor R, Plate I, Heinrich M, Michael M, Schöning R, Wittig H, Lutz-Bonengel S (2007) Mitochondrial D-loop (CA) n repeat length heteroplasmy: frequency in a German population sample and inheritance studies in two pedigrees. Int J Legal Med 121(3):207–213
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Thongngam P, Leewattanapasuk W, Bhoopat T, Sangthong P (2016) Nucleotide sequence analysis of the hypervariable region III of mitochondrial DNA in Thais. J Forensic Legal Med 41:10–14
Wilson MR, DiZinno JA, Polanskey D, Replogle J, Budowle B (1995) Validation of mitochondrial DNA sequencing for forensic casework analysis. Int J Legal Med 108:68–74
Acknowledgements
We acknowledge the Director, Anthropological Survey of India, and also express our gratitude to Dr. C. R. Satyanarayanan, Deputy Director and Head of Office, Anthropological Survey of India, SRC, Mysore. The authors also thank the Chairperson, Department of Studies in Zoology, University of Mysore.
Availability of data and materials
Reported variable sequences in this article are deposited in the GenBank under accession numbers MF959506 - MF959509.
Author information
Authors and Affiliations
Contributions
CS, AC, and JSR contributed to the analysis and elucidation of the data. JSR supervised the study. CS, JSR, and MSK drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The Institutional Ethical Committee of the Anthropological Survey of India and University of Mysore approved the protocol and ethical clearance of the study. Written informed consent was obtained from all the study individuals.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Sylvester, C., Krishna, M.S., Rao, J.S. et al. Allele frequencies of mitochondrial DNA HVR III 514–524 (CA)n dinucleotide repeats in the Urali Kuruman tribal population of South India. Egypt J Forensic Sci 8, 52 (2018). https://doi.org/10.1186/s41935-018-0083-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41935-018-0083-5