Abstract
Background
Human malaria is a major threat in rural communities of central Vietnam. Anopheles dirus and Anopheles minimus species are critical malaria vectors in Vietnam, which transmit Plasmodium parasites. However, the entomological aspects of malaria transmission in some of the central provinces of Vietnam remain unexplored. Hence, a cross-sectional entomological survey was carried out to identify the malaria vector species and the transmission of Plasmodium parasites in seven endemic provinces of Vietnam.
Methods
Mosquitoes were collected from seven provinces, Gia Lai, Khanh Hoa, Phu Yen, Ninh Thuan, Binh Thuan, Dong Nai, and Binh Phuoc. The collection was conducted for four to eight consecutive nights using three established methods, indoor and outdoor human landing catches and light trap method. Nested-PCR analysis was performed to detect the Plasmodium species in the separated thorax and the abdomen of the individual mosquitoes.
Results
A total of 2278 mosquitoes belonging to one of the four species of anopheline mosquitoes, An. dirus, An. maculatus, An. aconitus, and An. minimus were collected. Among the collected mosquitoes, 1398 were analysed using nested-PCR, of which, 40 mosquitoes were positive for Plasmodium parasites. Most of these parasites were detected in the samples from the thorax region, followed by the abdominal portion. The parasites were detected in both the thorax and abdomen of An. dirus. Seven species of Plasmodium parasites were detected during the analysis, of which, Plasmodium inui was the most common species, followed by Plasmodium falciparum, Plasmodium vivax, Plasmodium cynomolgi, Plasmodium coatneyi, Plasmodium knowlesi, and Plasmodium fieldi. Out of the 49 positive samples, 12 showed mixed infections. Co-infection of P. inui with human and other non-human primate Plasmodium species was common.
Conclusions
This study demonstrated the presence of human and non-human primate Plasmodium infection in An. dirus, a predominant malarial vector. Further, we showed that An. maculatus and An. minimus species also take part in malarial transmission. This might potentially lead to an alarming situation conducive for the emergence of novel zoonotic malaria.
Similar content being viewed by others
Background
Malaria is the most important infection caused by protozoans in humans. “Forest malaria” is a term frequently used to describe malaria prevalence in Southeast Asia. Transmission of Plasmodium parasites have been extensively found in the forest areas of southern and central Vietnam [1, 2]. Plasmodium knowlesi, a primate malarial parasite, has been detected in both human and Anopheles dirus in the central region of Vietnam [1, 3,4,5,6]. Furthermore, An. dirus from the Khanh Phu Commune, Khanh Vinh District, Khanh Hoa Province, was positive for both human and non-human primate Plasmodium parasites, of which, Plasmodium vivax was the most common species, followed by P. knowlesi, P. inui, P. cynomolgi, P. coatneyi, and P. falciparum [3, 4]. In few provinces of Southern Vietnam, residents have difficulty accessing malaria prevention services, especially the vector control measures, because of the local cultivation habits associated with sleeping in the forest and farm huts. These human behaviours and several environmental factors contribute to the risk of sylvatic transmission of Plasmodium parasites leading to zoonotic malarial infection among these people in Southern Vietnam [1, 6]. Since malaria control is quite unsuccessful in remote areas such as mountainous and forest areas, malaria continues to be an important health problem [1, 2, 7, 8].
The present study focuses on the determination of the various malaria vector species and their role in malaria transmission. We also demonstrated the prevalence of Plasmodium parasite infection in the vector species in seven provinces, Gia Lai, Khanh Hoa, Phu Yen, Ninh Thuan, Binh Thuan, Dong Nai, and Binh Phuoc of Vietnam. Hence, a detailed analysis of the mosquitoes and Plasmodium parasites infected vectors in these areas are necessary to assess the risk of potential zoonotic infection with non-human primate parasites in Southern Vietnam.
Materials and methods
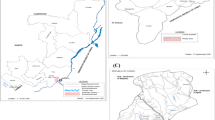
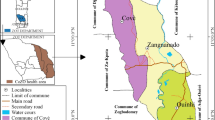
Mosquitoes were collected in 10 communes from 7 provinces (Fig. 1); Chu Rcam (13°30′ N; 108°60′ E) and Ia R Sai (13°33′ N; 108°62′ E) communes, Krong Pa District, Gia Lai Province; Ea Charang (13o3′ N; 108o52′ E) Commune, Son Hoa District, Phu Yen Province; Son Thai (12°24′ N; 108°81′ E) and Khanh Thuong (12°28′ N; 108°77′ E) communes, Khanh Vinh District, Khanh Hoa Province; Phuoc Binh (11o57′ N;108o45′ E) Commune, Bac Ai District, Ninh Thuan Province; Phan Tien (11o18′ N; 108o10′ E) Commune, Bac Binh District, Binh Thuan Province; Hieu Liem (11o10′52′′ N; 106o57′ E) Commune, Vinh Cưu District, Dong Nai Province; and Bu Gia Map Commune (11o56′ N; 106 o59′ E), Binh Phuoc Province; at the beginning of the rainy season (May) and the beginning of the dry season (December) between 2005 and 2018. However, as rainy and dry seasons commence early in Gia Lai, Dong Nai, and Binh Phuoc provinces, mosquito collection was carried out a month prior. Mosquitoes were collected at the hamlet and farm hut sites within the study area. The hamlet sites are small settlements away from the forest, with people living in one-storied, brick or wooden houses and using mosquito nets. The farm hut sites are located at the forest fringe, with simple huts located at a distance from each other and no electricity. Some people at such sites do not use mosquito nets. Moreover, these areas were previously reported to be hyper- to holo-endemic for malarial infections.
Mosquito collection was conducted for four to eight consecutive nights using three different collection methods—indoor human landing catches (IHLC), outdoor human landing catches (OHLC), and indoor light traps (ILT). IHLC and OHLC methods were conducted in the field from 18:00 to 24:00 h. Mosquitoes were morphologically distinguished in the field using a standardised method to identify anopheline mosquitoes in Southeast Asia [9]. Mosquitoes were stored in small tubes over silica gel. The tubes were labelled individually by collected site, collected time, and the collection methods for subsequent analyses. This study was reviewed and approved by the research ethics committee of the National Institute of Malariology, Parasitology, and Entomology, Ministry of Health, Vietnam (366/QD118-VSR) and by the ethics committee of Fujita Health University, Japan (HM17-050). People involved in the collection process were informed about the objectives, processes, and procedures of this study. Oral informed consent was obtained from them, and they were regularly screened for malaria for 1 month and in case of infection, they were promptly treated with artemisinin-based combination therapy (ACT).
DNA was extracted from the thoracic and abdominal portions of the mosquitoes, and subsequent PCR analysis was carried out as previously described elsewhere [3]. The dissection instruments were sterilised using alcohol and flame to prevent contamination. Genomic DNA was extracted using REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich, MO, USA). Nested-PCR assays to specifically identify human and non-human primate Plasmodium species were performed as described previously [10, 11]. The genus-specific primers, rPLU-1/rPLU-5, were used in the primary amplification (nest 1) and performed as described earlier. Detection of species-specific 18S rRNA (nest 2) was performed as previously described [10, 11]. For nest 2, 2 μL of 50× nest 1 amplification product (diluted with PCR grade water) was used as the template. The PCR products were separated by electrophoresis on 1.5% agarose gels and visualised after staining with ethidium bromide. The DNA bands were analysed using Lane and Spot Analyzer software (Atto, Tokyo, Japan). To confirm the P. knowlesi infection, circumsporozoite protein (CSP) gene was detected as previously described [12]. Primer sequences for 18S rRNA of human malaria parasites and primers to amplify CSP gene of P. knowlesi were as previously described [10,11,12]. A sample was considered positive for P. knowlesi only if both the assays (Kn1f/Kn3r and PkCSP) give positive results.
The 18S rRNA PCR amplicons of the non-human primate Plasmodium species were subjected to PCR clean-up using Wizard SV Gel and PCR Clean-up System (Promega, Tokyo, Japan) according to the manufacturer’s instructions. These products were then used for sequencing analysis with the BigDye Terminator v3.1 Cycle Sequencing Premix Kit (Applied Biosystems, Inc. Tokyo, Japan). The sequencing products were run on an ABI/Hitachi 3130 × 1 Genetic Analyzer (ABI), and the resultant nucleotide sequences were analysed using software Genetyx version 11 (Genetyx Corporation, Tokyo, Japan).
Results and discussion
In the present study, we focused on four Anopheles species—An. dirus, An. minimus, An. maculatus, and An. aconitus—as they are regarded as the primary and secondary malaria vectors in Vietnam. In total, 2278 mosquitoes belonging to the four Anopheles species were collected during the survey in the study provinces (Table 1). Number of collected mosquitoes in Khanh Hoa, Gia Lai, Phu Yen, Ninh Thuan, Binh Thuan, Dong Nai, and Binh Phuoc provinces were 332, 433, 63, 8, 437, 32, and 573, respectively. Among the four collected Anopheles species, An. dirus was the most common species (68%), followed by An. maculatus (21%), An. aconitus (8%), and An. minimus (3%) (Table 1). An. dirus and An. minimus, the critical vectors for Plasmodium parasites, were mainly collected by OHLC method (73%), followed by ILT (19%) and IHLC methods (8%) (Additional file 1). These results are because of the difference in environmental factors such as houses and differences in human behaviour depending on the collection site. [13,14,15]. Moreover, such differences can also be because of differences in biting behaviour of malaria vector mosquitoes [14,15,16,17].
In this study, 1386 mosquitoes randomly selected among the collected mosquitoes were subjected to nested PCR analysis for the detection of Plasmodium parasites (Table 2). The analysis indicated that 40 mosquitoes were positive for Plasmodium DNA. Among the 40 PCR-positive mosquitoes, the samples positive for Plasmodium DNA in the thorax, abdomen, and both regions were 20, 14, and 6, respectively (Additional file 2). From the sample collection data, we observed that 12/288 were positive in Khanh Hoa Province, 1/264 in Gia Lai Province, 3/133 in Phu Yen Province, 1/8 in Ninh Thuan Province, 1/32 in Dong Nai Province, and 23/573 in Binh Phuoc Province. No samples were tested positive in Binh Thuan Province (0/100) (Table 2). These PCR-positive samples were detected in An. dirus (37/1097), An. minimus (2/68), and An. maculatus (1/137) but not in An. aconitus (0/96). These mosquitoes were collected in the fields by ILT and OHLC methods but not by IHLC method (Additional file 3). These results were suggested to be dependent on differences in environmental factors, human behaviour, and mosquito behaviour [13,14,15,16,17].
Using species-specific PCR analysis, seven Plasmodium species were detected from 49 mosquito samples consisting of 28 samples of the thorax and 21 samples of the abdomen. Among the 49 PCR-positive samples, P. inui was the most common species (19/49), followed by P. falciparum (18/49), P. vivax (14/49), P. cynomolgi (9/49), P. coatneyi (3/49), P. fieldi (1/49), and P. knowlesi (1/49) (Table 3). Among these samples, mono-infection of six Plasmodium species was detected. Number of samples with mono-infection positive for P. falciparum, P. vivax, P. inui, P. cynomolgi, P. coatneyi, and P. fieldi were 14, 5, 10, 6, 1, and 1, respectively. Mixed infections with two or three Plasmodium species were detected in 12 samples. We observed that the co-infection of P. inui or P. vivax with human and non-human primate Plasmodium species was the most common, followed by P. falciparum co-infection with human and non-human primate Plasmodium parasites (Table 3). In addition, we also observed that there was no significant change in the Plasmodium infection rate with respect to its presence in the thorax or abdomen of the vector.
From previous literature, it is known that at least 30 of about 450 Anopheles species are known vectors of human and non-human primate Plasmodium parasites [3, 18]. In the Southeast Asian countries, An. dirus, An. minimus, An. maculatus, An. epiroticus, An. jeyporiensis, An. aconitus, An. sinensis, and An. sundaicus are the common vectors of Plasmodium parasites [2, 13, 19,20,21]. In the present study, four Anopheles species were collected from 10 different sites in Vietnam using 3 established collection methods. Previous studies demonstrated An. dirus and An. minimus to be the primary malaria vectors. An. dirus is considered the most dangerous malaria vector in Vietnam at present. An. maculatus and An. aconitus are the known secondary vectors for malarial infections [2,3,4,5, 13]. Among the mosquitoes collected in this study, An. dirus was the dominant vector species, whereas, An. minimus was present in small number. Previous long-term malaria project in Khanh Phu Commune near the Son Thai and Khanh Thuong communes of Khanh Vinh District, Khanh Hoa Province, reported that An. minimus was the major vector mosquito. But recently, An. dirus has emerged to be the dominant species [13]. Changes in the dominant species of the vector mosquitoes could be attributed to the environmental changes, modernisation, and the mosquito response after a prolonged exposure to insecticides used in malarial control [13, 15, 22].
In this study, two species of human parasites, P. falciparum and P. vivax and five species of non-human primate parasites, P. inui, P. cynomolgi, P. coatneyi, P. knowlesi, and P. fieldi were identified from the thorax and abdomen of the vector mosquitoes. Approximately, one-fourth of the vector mosquitoes were infected with two or three Plasmodium species, often in combination of human and non-human primate parasites. Similar findings were reported in a previous study in Khanh Phu Commune [3]. All these parasites are transmitted by anopheline mosquito vectors, which not only naturally infect macaques but also bite humans to cause malaria. These results indicated that these areas are at the risk of infection with both human and non-human primate Plasmodium parasites. These results were found to be applicable not only in a limited area but also in a wide region, further emphasising the potential risks involved in the outbreak of massive zoonotic malarial attack.
Conclusions
The dominant species transmitting the Plasmodium parasites in the provinces of Vietnam chosen for this study was An. dirus. By PCR analysis, seven species of Plasmodium parasites, P. inui, P. falciparum, P. vivax, P. cynomolgi, P. coatneyi, P. knowlesi, and P. fieldi were detected in the vectors. Twelve out of 49 PCR-positive samples showed mixed species infection, most of which were a co-infection of P. inui with P. vivax or non-human primate Plasmodium species. These results suggest that humans are at a risk for infection with both human and non-human primate Plasmodium parasites, leading to a situation conducive for the emergence of novel zoonotic malaria in Southern Vietnam.
References
Marchand RP, Culleton R, Maeno Y, Quang NT, Nakazawa S. Co-infections of Plasmodium knowlesi, P. falciparum, and P. vivax among humans and Anopheles dirus mosquitoes, southern Vietnam. Emerg Infect Dis. 2011. https://doi.org/10.3201/eid1707.101551.
Grietens KP, Xuan XN, Ribera J, Duc TN, Wv B, Ba NT, Van KP, Xuan HL, D'Alessandro U, Erhart A. Social determinants of long lasting insecticidal hammock use among the Ra-glai ethnic minority in Vietnam: implications for forest malaria control. PLoS One. 2012. https://doi.org/10.1371/journal.pone.0029991.
Maeno Y, Quang NT, Culleton R, Kawai S, Masuda G, Nakazawa S, Marchand RP. Humans frequently exposed to a range of non-human primate malaria parasite species through the bites of Anopheles dirus mosquitoes in South-Central Vietnam. Parasit Vectors. 2015. https://doi.org/10.1186/s13071-015-0995-y.
Maeno Y. Molecular epidemiology of mosquitoes for the transmission of forest malaria in south-Central Vietnam. Trop Med Health. 2017. https://doi.org/10.1186/s41182-017-0065-6.
Nakazawa S, Marchand RP, Quang NT, Culleton R, Manh ND, Maeno Y. Anopheles dirus co-infection with human and monkey malaria parasites in Vietnam. Int J Parasitol. 2009. https://doi.org/10.1016/j.ijpara.2009.08.005.
Pongvongsa T, Culleton R, Ha H, Thanh L, Phongmany P, Marchand RP, Kawai S, Moji K, Nakazawa S, Maeno Y. Human infection with Plasmodium knowlesi on the Laos-Vietnam border. Trop Med Health. 2018. https://doi.org/10.1186/s41182-018-0116-7.
Pongvongsa T, Ha H, Thanh L, Marchand R, Nonaka D, Tojo B, Phongmany P, Moji K, Kobayashi J. Joint malaria survey lead toward improved cross-border cooperation between Savannakhet province, Laos and Quang Tri province, Vietnam. Malar J. 2012. https://doi.org/10.1186/1475-2875-11-262.
Fornace KM, Herman LS, Abidin TR, Chua TH, Daim S, Lorenzo PJ, Grignard L, Nuin NA, Ying LT, Grigg MJ, William T, Espino F, Cox J, Tetteh KKA, Drakeley CJ. Exposure and infection to Plasmodium knowlesi in case study communities in northern Sabah, Malaysia and Palawan, The Philippines. PLoS Negl Trop Dis. 2018. https://doi.org/10.1371/journal.pntd.0006432.
Hung NM, De VK, Chinh VD, Manh ND, Trung HD, Duy BL. Key to Anopheline mosquitoes in Vietnam. Hanoi: Medical Publishing house; 2008.
Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999. https://doi.org/10.4269/ajtmh.1999.60.687.
Lee KS, Divis PC, Zakaria SK, Matusop A, Julin RA, Conway DJ, Cox-Singh J, Singh B. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011. https://doi.org/10.1371/journal.ppat.1002015.
Vythilingam I, Noorazian YM, Huat TC, Jiram AI, Yusri YM, Azahari AH, Norparina I, Noorrain A, Lokmanhakim S. Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia. Parasit Vectors. 2008. https://doi.org/10.1186/1756-3305-1-26.
Hii J, Rueda LM. Malaria vectors in the greater Mekong subregion: overview of malaria vectors and remaining challenges. Southeast Asian J Trop Med Public Health. 2013;44(Suppl 1):73–165 discussion 306-7.
Ngo CT, Dubois G, Sinou V, Parzy D, Le HQ, Harbach RE, Manguin S. Diversity of Anopheles mosquitoes in Binh Phuoc and Dak nong provinces of Vietnam and their relation to disease. Parasit Vectors. 2014. https://doi.org/10.1186/1756-3305-7-316.
Van Bortel W, Chinh VD, Berkvens D, Speybroeck N, Trung HD, Coosemans M. Impact of insecticide-treated nets on wild pyrethroid resistant Anopheles epiroticus population from southern Vietnam tested in experimental huts. Malar J. 2009. https://doi.org/10.1186/1475-2875-8-248.
Van Bortel W, Trung HD, Hoi le X, Van Ham N, Van Chut N, Luu ND, Roelants P, Denis L, Speybroeck N, D'Alessandro U, Coosemans M. Malaria transmission and vector behaviour in a forested malaria focus in Central Vietnam and the implications for vector control. Malar J. 2010. https://doi.org/10.1186/1475-2875-9-373.
Sanh NH, Van Dung N, Thanh NX, Trung TN, Van Co T, Cooper RD. Forest malaria in central Vietnam. Am J Trop Med Hyg. 2008;79:652–4.
White GB. Malaria vector ecology and genetics. Br Med Bull. 1982. https://doi.org/10.1093/oxfordjournals.bmb.a071760.
Trung HD, Van Bortel W, Sochantha T, Keokenchanh K, Quang NT, Cong LD, Coosemans M. Malaria transmission and major malaria vectors in different geographical areas of Southeast Asia. Tropical Med Int Health. 2004. https://doi.org/10.1046/j.1365-3156.2003.01179.x.
Trung HD, Bortel WV, Sochantha T, Keokenchanh K, Briët OJ, Coosemans M. Behavioural heterogeneity of Anopheles species in ecologically different localities in Southeast Asia: a challenge for vector control. Tropical Med Int Health. 2005. https://doi.org/10.1111/j.1365-3156.2004.01378.x.
Klein TA, Harrison BA, Dixon SV, Burge JR. Comparative susceptibility of Southeast Asian Anopheles mosquitoes to the simian malaria parasite Plasmodium cynomolgi. J Am Mosq Control Assoc. 1991;7:481–7.
Cui L, Yan G, Sattabongkot J, Cao Y, Chen B, Chen X, Fan Q, Fang Q, Jongwutiwes S, B, Xu J, Zheng B, Zhong D, Zhou G. Malaria in the Greater Mekong Subregion: heterogeneity and complexity. Acta Trop 2012. https://doi.org/10.1016/j.actatropica.2011.02.016.
Acknowledgements
We would like to thank Kaoru Hori for her technical support. We appreciate the support of the staffs from the seven provinces, Gia Lai, Khanh Hoa, Phu Yen, Ninh Thuan, Binh Thuan, Dong Nai, and Binh Phuoc and the local authorities.
Funding
This study was partly supported by the following grants- (i) WHO, Vietnam, (ii) JSPS KAKENHI Grant Numbers- 26360029 and 17H04513, (iii) the Cooperative Research Grants from Joint Usage/Research Centre on Tropical Disease, Institute of Tropical Medicine, Nagasaki University (2015-Ippan-14, 2016-Ippan-9, 2017-Ippan-2, 2018-Ippan-7).
Availability of data and materials
The data supporting the conclusions of this article are included in the article.
Author information
Authors and Affiliations
Contributions
VDC, VVH, GM, and YM contributed to the study design and conception. VDC, VVH, GM, HT, SK, TA, and YM helped in the fieldwork and data collection. VDC, VVH, GM, HT, SK, TA, and YM carried out the experiments and prepared the manuscript. VDC and VVH helped in the statistical analyses. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was reviewed and approved by the research ethics committee of the National Institute of Malariology, Parasitology and Entomology, Ministry of Health, Vietnam (366/QD-VSR) and the ethics committee of Fujita Health University, Japan (HM17-050). The people involved in mosquito collection and householders were informed about the objectives, processes, and procedures of this study. Oral informed consents were obtained from them.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Number of mosquitoes collected using different collection methods; outdoor human landing catches (OHLC), indoor human landing catches (IHLC), and indoor light traps (ILT). (DOCX 16 kb)
Additional file 2:
Results of the detected Plasmodium spp. infection in the thorax and abdomen of the collected mosquitoes from the study sites. (DOCX 17 kb)
Additional file 3:
Results of the detected Plasmodium spp. infection by collection method, outdoor human landing catches (OHLC), indoor human landing catches (IHLC), and indoor light traps (ILT). (DOCX 18 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chinh, V.D., Masuda, G., Hung, V.V. et al. Prevalence of human and non-human primate Plasmodium parasites in anopheline mosquitoes: a cross-sectional epidemiological study in Southern Vietnam. Trop Med Health 47, 9 (2019). https://doi.org/10.1186/s41182-019-0139-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41182-019-0139-8