Abstract
Background
Dissimilarity in community composition is one of the most fundamental and conspicuous features by which different forest ecosystems may be distinguished. Traditional estimates of community dissimilarity are based on differences in species incidence or abundance (e.g. the Jaccard, Sørensen, and Bray-Curtis dissimilarity indices). However, community dissimilarity is not only affected by differences in species incidence or abundance, but also by biological heterogeneities among species.
Methods
The objective of this study is to present a new measure of dissimilarity involving the biological heterogeneity among species. The “discriminating Avalanche” introduced in this study, is based on the taxonomic dissimilarity between tree species. The application is demonstrated using observations from five stem-mapped forest plots in China and Mexico. We compared three traditional community dissimilarity indices (Jaccard, Sørensen, and Bray-Curtis) with the “discriminating Avalanche” index, which incorporates information, not only about species frequencies, but also about their taxonomic hierarchies.
Results
Different patterns emerged for different measures of community dissimilarity. Compared with the traditional approaches, the discriminating Avalanche values showed a more realistic estimate of community dissimilarities, indicating a greater similarity among communities when species were closely related.
Conclusions
Traditional approaches for assessing community dissimilarity disregard the taxonomic hierarchy. In the traditional analysis, the dissimilarity between Pinus cooperi and Pinus durangensis would be the same as the dissimilarity between P. cooperi and Arbutus arizonica. The dissimilarity Avalanche dissimilarity between P. cooperi and P. durangensis is considerably lower than the dissimilarity between P. cooperi and A. arizonica, because the taxonomic hierarchies are incorporated. Therefore, the discriminating Avalanche is a more realistic measure of community dissimilarity. This main result of our study may contribute to improved characterization of community dissimilarities.
Similar content being viewed by others
Background
Dissimilarity in community composition is one of the most conspicuous features of forest ecosystems (Jost et al. 2011). Assessing compositional differences between forest communities is an important issue for several reasons. Dissimilarities between forest communities can reveal certain mechanisms that generate and maintain forest biodiversity and specific habitat effects that shape forest composition and structure (Socolar et al. 2016). Assessing dissimilarities between forest communities is essential for evaluating species invasions, changes caused by selective tree harvesting, or effects of climate change on species composition. In addition, effective measures of community dissimilarity may contribute to more meaningful classifications of forest vegetation (Wehenkel et al. 2014).
Jaccard (1900) was probably the first who proposed a method for measuring the degree of community similarity and dissimilarity based on the number of species shared by two communities and the number of species unique to each of them. Two additional indices were subsequently proposed to estimate the difference between communities (Sørensen 1948; Bray and Curtis 1957). The Jaccard and Sørensen indices are using species presence/absence data. The Bray-Curtis index, a modified version of the Sørensen index, includes species abundances (Chao et al. 2005). These three indices have become the most widely used measures for assessing community similarity or dissimilarity in community ecology (Anderson et al. 2006). In addition, several other species incidence- or abundance-based indices have been developed (Chao et al. 2005; Legendre and Legendre 2012). Examples are the Chi-square distance index (Fenelon and Lebart 1971), the Canberra index (Lance and Williams 1967; Stephenson et al. 1972), and the Morisita-Horn index (Magurran 2004).
These indices have been widely used in forest ecology, and they contribute substantially to the understanding of community dissimilarity. However, community dissimilarity is not only affected by differences in species abundance or incidence, but also by the biological heterogeneity among species (Clarke and Warwick 1998, 2001). Accordingly, the purpose of this study is to evaluate a new measure of community dissimilarity that incorporates both the information of species frequencies and the biological heterogeneity among species. The new approach is based on the “Avalanche” index proposed by Ganeshaiah et al. (1997) and Ganeshaiah and Shaanker (2000). The biological dissimilarity among species can be calculated using species taxonomic, genetic or morphometric information.
Data and methods
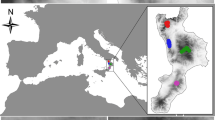
We are using observations from five 1-ha (100 m × 100 m) forest plots, three from Mexico and two from China, to demonstrate the new approach (Fig. 1).
Observational field plots
The three Mexican plots are located in the communal forests of Durango (22°20′–26°47′ N; 103°46′–107°12′ W), which occupy about 23% of the area of Sierra Madre Occidental. The elevation above sea level varies between 363 and 3200 m (average 2264 m). The precipitation ranges from 443 to 1452 mm, with an annual average of 917 mm, while the mean annual temperature varies from 8.2 to 26.2 °C, with an annual average of 13.3 °C (González-Elizondo et al. 2012; Silva-Flores et al. 2014). The predominant forest types are uneven-aged, semi-natural forests, which are dominated by Pinus spp. and Quercus spp., and often in mixture with Arbutus spp., Juniperus spp., and Pseudotsuga menziesii (Silva-Flores et al. 2014; Lujan-Soto et al. 2015). In Durango, many local residents depend on the forests for their livelihood. But despite the high biological, cultural and socio-economic importance of the Sierra Madre Occidental, these forests are not very well known. Details about the history of these unique ecosystems may be found in Burgos and Villa (1974), and Corral-Rivas et al. (2015).
The three forests where the plots are located, have been managed selectively by local communities known as Ejidos. Previous commercial harvests were based on maintaining an inverse J-shaped diameter class distribution (Virgilietti and Buongiorno 1997). The sites have been protected for many years, and are located in the vicinity of a National Park. The exact treatment histories are not known. The permanent field plots, named after the local Ejidos “La Victoria”, “San Esteban” and “Mil Diez”, are located in the region of El Salto, Pueblo Nuevo, Durango. All the woody stems in the three plots with DBH ≥ 5 cm were identified, measured and stem-mapped. A total of 2041 individual trees belonging to eleven species, four genera, and four families are included in the three plots. The main species in terms of basal area are Pinus cooperi, Quercus sideroxyla, and Pinus durangensis.
The two plots from China are from unmanaged forests. One plot is located in a temperate broad-leaved Korean pine (Pinus koraiensis) mixed forest in Jiaohe Forest Experimental Zone (43°51′–44°05′ N, 127°35′–127°51′ E), in Jilin Province, northeastern China (hereafter referred to as “Jiaohe”). The other plot is situated in a subtropical evergreen broad-leaved forest in Jiulian Mountain National Natural Reserve (24°29′–24°39′ N; 114°22′–114°32′ E), in Jiangxi Province, southeastern China (hereafter “Jiulian”). The mean annual temperature in Jiaohe is 3.8 °C, with average monthly temperature ranges from − 18.6 °C to 21.7 °C, while the mean annual temperature in Jiulian is 17.4 °C, with average monthly temperature ranges from 6.8 °C to 24.4 °C. The mean annual precipitation is 696 mm (Jiaohe) and 2156 mm (Jiulian), respectively (Hao et al. 2018). There are 675 individuals belonging to 28 species, 18 genera and 13 families in Jiaohe plots, and 1060 individuals belonging to 112 species, 68 genera and 38 families in the Jiulian plots. Details of the five research plots are presented in Table 1.
Dissimilarity between species
The biological dissimilarity (or “distance”) between species refers to the difference between species in terms of certain genetic or morphological characteristics (Faith 1992; Ganeshaiah et al. 1997; Clarke and Warwick 1998). The biological dissimilarity can be assessed based on plant traits (Ganeshaiah et al. 1997), phylogeny (Faith 1992), or taxonomy (Clarke and Warwick 1998, 1999). In this study, we choose to calculate the biological dissimilarity between species using the information of a Linnean taxonomy. Measuring the taxonomic distance is relatively straightforward, compared with the use of more complex plant traits or phylogeny. The taxonomic distances are estimated based on a table of classification which includes the five taxonomic levels (species, genus, family, order, and group).
The standard botanical nomenclature follows The Plant List (TPL, www.theplantlist.org), which represents an internationally accepted standard database for plant nomenclature. The taxonomic information is extracted from the APG IV classification system (Angiosperm Phylogeny Group 2016) and Christenhusz et al. (2011), for Angiosperms and Gymnosperms, respectively. The distances between species are scaled such that the longest path length between taxa is 1. For example, the distance would be 1 if two individuals belong to different taxonomic groups (Angiosperms and Gymnosperms). The distance would be 0.8 if two individuals belong to different orders, but share the same group (e.g. Pinales and Fagales). By that analogy, the distance would be 0.2 for two individuals that belong to different species but share the same genus. The normalized distances between two individuals that differ at different levels within the taxonomic hierarchy are presented in Table 2.
The Jaccard, Sørensen and Bray-Curtis indices
The Jaccard, Sørensen, and Bray-Curtis indices are presented in Table 3 for easy reference. These indices are well known and widely applied, especially in the assessment of community dissimilarity.
The simple, complete, discriminating and plain avalanche
The “Avalanche index” (Ganeshaiah et al. 1997; Ganeshaiah and Shaanker 2000) represents a generalization of the phylogenetic diversity index proposed by Faith (1992). Which broadens the distance component to allow for any quantitative information that is biologically informative (not just phylogeny), and that can measure the proximity of any given pair of species (Talents et al. 2005). If only species incidence data are available, the simple Avalanche index (sA) is defined as follows:
where n is the number of species and dij is the biological distance between species i and j. sA can be standardized by normalizing the dij and dividing the sA by n(n-1). The result is a value in the interval [0,1]. This is a great advantage when compared with the Shannon index (or Hill numbers) of species diversity. If species abundance data are available, the complete Avalanche (cA), which estimates the biodiversity within a community, can be used:
where n is the number of species and dij is the taxonomic, phylogenetic or trait distance between species i and j; pi and pj are the relative frequencies of species i and j in the community. In addition, pi and pj can also be the relative basal area or biomass or other weights of species i and j. Based on these original “Avalanches”, we present a new measure called “discriminating Avalanche”, which can be used to quantify the biological dissimilarity (or distance) between two communities. The discriminating Avalanche (dA) is based on species frequencies and some measure of biological distance among species:
where \( {\Delta}_i^{a,b} \) refers to the absolute difference between the frequencies of species i in plots a and b (\( {\Delta}_i^{a,b} \) =| \( {p}_i^a \)– \( {p}_i^b \) |, \( {p}_i^a \) and \( {p}_i^b \) are the relative frequencies of species i in plots a and b), and \( {\Delta}_j^{a,b} \) is the equivalent for species j. If the maximum dij is known, then the dij can be normalized by dividing the actual values of dij by the maximum value of the dij, and dA will thus assume values in the interval [0, 1]. If we disregard the biological distance among species (as in the case of Jaccard, Sørensen, and Bray-Curtis) but only emphasize the differences in species frequency, we obtain the “Plain Avalanche” (pA):
The Plain Avalanche distances are expected to be greater than the discriminating Avalanche distances in situations where many individuals differ by species but share the same genus or family.
Results
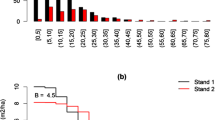
Based on the discriminating Avalanche (dA) the community distances among forests with species taxonomic information are obtained (Fig. 2). We simultaneously calculated the Jaccard, Sørensen, Bray-Curtis, and Plain Avalanche (pA) community dissimilarity indices, and found different patterns for different measures of dissimilarity (Fig. 3; Additional file 1: Table S1). Compared with the traditional measures, the discriminating Avalanche values showed relatively low distances, i.e. a greater similarity among plots, as closer relations among species are revealed by dA.
With regard to the three forest plots in Mexico, the Jaccard and Sørensen dissimilarities exhibit similar tendencies: a closer distance between plots San Esteban and Mil Diez, and a relatively greater distance between La Victoria and Mil Diez. The pA distances exhibit a tendency which is similar to Bray-Curtis: a closer distance between La Victoria and San Esteban, and a greater distance between San Esteban and Mil Diez. The dA distances are different from all others: a closer distance between La Victoria and San Esteban, and a greater distance between La Victoria and Mil Diez (Fig. 3; Additional file 1: Table S1). When there are no shared species between plots, the Jaccard, Sørensen, and Bray-Curtis distances are both 1, i.e. the maximum value of these indices. The pA and dA distances, although there are no shared species in the forest plots in China and Mexico, do not reach the maximum value. These results will be discussed in detail in the discussion section.
Discussion
Assessing the difference among communities has become a central issue in community ecology (Chao et al. 2005; Legendre and Cáceres 2013). In this study, we presented a new taxonomy-based approach, which is easy to implement, and which will bring new information compared to traditional approaches of assessing community dissimilarity, and can be applied flexibly in any pair of communities with any species richness.
Measures of community dissimilarity
Community “similarity” or “dissimilarity” is a qualitative human construct which has no precise mathematical definition. Nevertheless, measuring community dissimilarity should rely on some quantitative measure that can be devised for a specific purpose (Chao et al. 2005). Dissimilarity means “not the same”. When we state that two forest communities are the same, what we are really doing is to neglect those differences that we choose to ignore. Therefore, although numerous measures have been published, the development of new measures for quantifying community dissimilarity more accurately, more comprehensively and objectively remains a priority in community ecology (Chao et al. 2005).
One property that any new measure should have is that it should not be redundant with existing indices. Therefore, the new measure is compared with the well known Jaccard, Sørensen, and Bray-Curtis dissimilarity indices. The patterns of biological dissimilarity presented in Fig. 3 differ considerably. Based on these differences, the five measures of dissimilarity are assigned to three groups. The first group which we call the Species Presence-Absence group, includes the Jaccard and Sørensen dissimilarities. The Jaccard and Sørensen are based solely on species incidence (presence-absence) data, i.e. the number of species shared by two plots and the number of species unique to each. It is not surprising that Jaccard and Sørensen exhibit similar tendencies (e.g. a closer distance between plots San Esteban - Mil Diez, a relatively longer distance between plots La Victoria - Mil Diez, and the longest distance when two plots have no shared species). Six species are shared by San Esteban and Mil Diez, while only three species are unique to each of these two communities. The dissimilarity between the two plots is relatively small because the focus is only on species incidence. Jaccard calculates the unique (unshared) species as a proportion of the total number of species recorded in the two communities, while Sørensen gives double weight to the shared species (Table 3). The Sørensen dissimilarity is thus closely related to Jaccard, and always has a lower value than Jaccard.
The second group which we call the Species-Abundance group, includes the Bray-Curtis and Plain Avalanche dissimilarities. Both are calculated using the species abundance data. Bray-Curtis and Plain Avalanche dissimilarities exhibit a similar tendency (e.g. a closer distance between La Victoria - San Esteban, and a greater distance between San Esteban - Mil Diez). The results seem to contradict the Presence-Absence group. Although there are six species shared by plots San Esteban and Mil Diez, the difference in the species frequencies is great resulting in higher dissimilarities. The Bray-Curtis and Plain Avalanche dissimilarities are based on the same information: the absolute difference on species frequencies (Table 3). However, there are differences between them: Bray-Curtis is calculated as the sum of absolute difference for all species (\( \frac{\sum_{i=1}^n\mid {p}_i^a-{p}_i^b\mid }{\sum_{i=1}^n\left({p}_i^a+{p}_i^b\right)} \) or \( \frac{1}{2}\sum \limits_{j=i}^n\left({\Delta}_i^{a,b}\right) \), where \( {\Delta}_i^{a,b}=\mid {p}_i^a-{p}_i^b\mid \)), while the Plain Avalanche is calculated as the sum of products of pair-frequency differences for all species (\( \sum \limits_{i=1}^n\sum \limits_{j=1}^n\left({\Delta}_i^{a,b}{\Delta}_j^{a,b}\right) \)).
Compared with the Plain Avalanche, the discriminating Avalanche gives lower distances, i.e. a greater similarity among plots in these particular communities. This is due to the fact that in the Plain Avalanche, each species is treated as an entity that is different from another species. For example, the dissimilarity between P. cooperi and P. durangensis would be the same as the dissimilarity between P. cooperi and A. arizonica. However, in the discriminating Avalanche, the dissimilarity between P. cooperi and P. durangensis is considerably lower than the dissimilarity between P. cooperi and A. arizonica, because the taxonomic hierarchies are incorporated.
The maximum and minimum values of Jaccard, Sørensen, and Bray-Curtis are 1 (no shared species) and 0 (the same species composition), respectively. The minimum value of the discriminating Avalanche and the Plain Avalanche is 0 (the same species composition), while the maximum value is (\( 1-\frac{1}{n} \)), where n is the total number of species recorded in the two communities (n ≥ 2; refer to the the mathematics theorem inequality of arithmetic and geometric means). Therefore the maximum value depends on the number of species. For example, if there are 10 species recorded in the two communities, the maximum value will be (\( 1-\frac{1}{10}=0.9 \)); if there are 100 species, the maximum value will be (\( 1-\frac{1}{100}=0.99 \)). For an infinite number of species, the value would be ~ 1. The maximum value of dA is thus hard to be achieved in practice, even if there is no shared tree species betwee communities. We employed forest plots from China and Mexico to explain this property. In our example the forests in China and Mexico have almost no shared species. Jaccard, Sørensen, and Bray-Curtis therefore both give the maximum pairwise dissimilarities (Fig. 3). However, from a more evolutionary perspective, the regions are not totally and equally dissimilar and can be compared in terms of their relative taxonomic similarity. Therefore, the discriminating Avalanche is a more realistic measure of community dissimilarity.
Future work: estimating biological distances based on optimization
This study has shown that the Avalanche approach has the potential to be applied in discriminating among forest communities. Instead of using the Avalanche approach for assessing biological distances, the transportation model of linear programming may also be suitable for evaluating the biological distance between forest communities, based on the objective to minimize the “cost” of transforming community i into community j:
where the distanceij is some measure of the biological distance between species i and j. The Xij are the numbers (or relative proportions) of the different species in communities i and j. The constraints would be the numbers (or relative proportions) of the different species in communities i and j:
\( \sum \limits_{\mathrm{j}}^{\mathrm{n}}{\mathrm{X}}_{\mathrm{i}\mathrm{j}}\le {\mathrm{available}}_{\mathrm{i}} \) and \( \sum \limits_{\mathrm{i}}^{\mathrm{m}}{\mathrm{X}}_{\mathrm{i}\mathrm{j}}\ge {\mathrm{required}}_{\mathrm{j}} \) (6).
The potential of this approach will be evaluated in future studies.
Conclusions
This study presents a new method for estimating compositional dissimilarities (distances) between forest communities. Estimates of the biological “distance” are based on the Avalanche concept, using a simple taxonomic hierarchy and species frequencies. To increase the contrast and interpretation of this new approach, compositional dissimilarities of forest communities are also assessed using three more traditional approaches, the Jaccard, Sørensen, and Bray-Curtis community dissimilarity indices. The results suggest that the discriminating Avalanche approach is not redundant but complementary and possibly more meaningful than the existing approaches. Such “distance” estimates could reveal the degree of biological relatedness of forest ecosystems in different regions of the world, estimate the effects of habitat heterogeneity on community composition and diversity, and improve an assessment of the degree of species invasion or anthropogenic disturbance with reference to an assumed potential natural vegetation.
Availability of data and materials
Please contact the corresponding author for data requests.
Abbreviations
- APG IV:
-
Angiosperm phylogeny group IV
- cA:
-
Complete Avalanche
- dA:
-
Discriminating Avalanche
- DBH:
-
Diameter at breast height
- pA:
-
Plain Avalanche
- sA:
-
Simple Avalanche
- TPL:
-
The Plant List
References
Anderson MJ, Ellingsen KE, McArdle BH (2006) Multivariate dispersion as a measure of beta diversity. Ecol Lett 9(6):683–693
Angiosperm Phylogeny Group (2016) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc 181(1):1–20
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27(4):325–349
Burgos MF, Villa SAB (1974) La silvicultura en el desarrollo económico-social de México. UIEF San Rafael SAG-SFF México, DF Bol Núm 5:144
Chao A, Chazdon RL, Colwell RK, Shen TJ (2005) A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett 8(2):148–159
Christenhusz MJ, Reveal JL, Farjon A, Gardner MF, Mill RR, Chase MW (2011) A new classification and linear sequence of extant gymnosperms. Phytotaxa 19(1):55–70
Clarke KR, Warwick RM (1998) A taxonomic distinctness index and its statistical properties. J Appl Ecol 35(4):523–531
Clarke KR, Warwick RM (1999) The taxonomic distinctness measure of biodiversity: weighting of step lengths between hierarchical levels. Mar Ecol Prog Ser 184:21–29
Clarke KR, Warwick RM (2001) A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar Ecol Prog Ser 216:265–278
Corral-Rivas JJ, Hernández-Díaz JC, López-Sánchez CA, Luján-Soto JE, Gadow KV (2015) Ejido Borbollones, Durango, Mexico. Chapter 9. In: Siry JP, Bettinger P, Merry K, Grebner DL, Boston K, Ch C (eds) Forest Plans of North America, Academic Press, Elsevier, pp 61–68
Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biol Conserv 61(1):1–10
Fenelon JP, Lebart L (1971) Statistique et informatique appliquées. Dunod, Paris
Ganeshaiah KN, Chandrashekara K, Kumar ARV (1997) Avalanche index: a new measure of biodiversity based on biological heterogeneity of the communities. Curr Sci 73(2):128–133
Ganeshaiah KN, Shaanker RU (2000) Measuring biological heterogeneity of forest vegetation types: avalanche index as an estimate of biological diversity. Biodivers Conserv 9(7):953–963
González-Elizondo MS, González-Elizondo M, Tena-Flores JA, Ruacho-González L, López-Enríquez IL (2012) Vegetación de la sierra madre occidental, México: Una síntesis. Acta Bot Mex 100:351–403
Hao M, Zhang C, Zhao X, Gadow KV (2018) Functional and phylogenetic diversity determine woody productivity in a temperate forest. Ecol Evol 8:2395–2406
Jaccard P (1900) Contribution au problème de l’immigration post-glaciaire de la flore alpine. Bulletin de la Société Vaudoise des Sciences Naturelles 36:87–130
Jost L, Chao A, Chazdon RL (2011) Compositional similarity and β (beta) diversity. In: Magurran AE, McGill BJ (eds) Biological diversity: frontiers in measurement and assessment. Oxford University Press, New York, pp 66–84
Lance GN, Williams WT (1967) Mixed-data classificatory programs I - agglomerative systems. Austr Comp J 1(1):15–20
Legendre P, Cáceres MD (2013) Beta diversity as the variance of community data: dissimilarity coefficients and partitioning. Ecol Lett 16(8):951–963
Legendre P, Legendre L (2012) Numerical Ecology, 3rd edn. Elsevier, Amsterdam
Lujan-Soto JE, Corral-Rivas JJ, Aguirre-Calderón OA, Gadow KV (2015) Grouping forest tree species on the Sierra Madre occidental, Mexico. Allgemeine Forst und Jagdzeitung 186(3–4):63–71
Magurran AE (2004) Measuring biological diversity. Blackwell, Oxford
Silva-Flores R, Pérez-Verdín G, Wehenkel C (2014) Patterns of tree species diversity in relation to climatic factors on the Sierra Madre occidental, Mexico. PLoS One 9(8):e105034
Socolar JB, Gilroy JJ, Kunin WE, Edwards DP (2016) How should beta-diversity inform biodiversity conservation? Trend Ecol Evol 31(1):67–80
Sørensen TA (1948) A method of establishing groups of equal amplitude in plant sociology based on similarity of species content, and its application to analysis of the vegetation on Danish commons. Biologiske Skrifter Kongelige Danske Videnskabernes Selskab 5:1–34
Stephenson W, Williams WT, Cook SD (1972) Computer analyses of Petersen's original data on bottom communities. Ecol Monogr 42(4):387–415
Talents LA, Lovett JC, Hall JB, Hamilton AC (2005) Phylogenetic diversity of forest trees in the Usambara mountains of Tanzania: correlations with altitude. Bot J Linn Soc 149(2):217–228
Virgilietti P, Buongiorno J (1997) Modeling forest growth with management data: a matrix approach for the Italian Alps. Silv Fenn 31(1):27–42
Wehenkel C, Corral-Rivas JJ, Gadow KV (2014) Quantifying differences between ecosystems with particular reference to selection forests in Durango/Mexico. For Ecol Manag 316:117–124
Acknowledgements
Not applicable.
Funding
This study was financed by the Program of National Natural Science Foundation of China (31670643), the Key Project of National Key Research and Development Plan of China (2017YFC0504104), Beijing Forestry University Outstanding Young Talent Cultivation Project (2019JQ03001), and the National Forestry Commission (CONAFOR) of Mexico through the PRONAFOR program.
Author information
Authors and Affiliations
Contributions
MH, CZ and KG: designed the study, performed data analyses and wrote the manuscript; JJCR, MGNM, CZ and XZ: created the database of forest plots; MSGE and KNG: provided comments and other technical support. All authors discussed the results and commented on the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional file
Additional file 1:
Table S1. Pair distances between the five forest plots in China and Mexico, based on different measures of compositional dissimilarity between communities (DOCX 21 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hao, M., Corral-Rivas, J.J., González-Elizondo, M.S. et al. Assessing biological dissimilarities between five forest communities. For. Ecosyst. 6, 30 (2019). https://doi.org/10.1186/s40663-019-0188-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40663-019-0188-9