Abstract
Background
Meiotic recombination is an important source of genetic variability. Studies on mammals demonstrate a substantial interspecies variation in overall recombination rate, which is dependent mainly on chromosome (2n) and chromosome arm number (FN). Bird karyotypes are very conservative with 2n being about 78–82 and FN being 80–90 in most species. However, some families such as Apodidae (swifts) and Falconidae (falcons) show a substantial karyotypic variation. In this study, we describe the somatic and pachytene karyotypes of the male Common Swift (Apus apus) and the pachytene karyotype of the male Eurasian Hobby (Falco subbuteo) and estimate the overall number and distribution of recombination events along the chromosomes of these species.
Methods
The somatic karyotype was examined in bone marrow cells. Pachytene chromosome spreads were prepared from spermatocytes of adult males. Synaptonemal complexes and mature recombination nodules were visualized with antibodies to SYCP3 and MLH1 proteins correspondingly.
Results
The karyotype of the Common Swift consists of three metacentric, three submetacentric and two telocentric macrochromosomes and 31 telocentric microchromosomes (2n = 78; FN = 90). It differs from the karyotypes of related Apodidae species described previously. The karyotype of the Eurasian Hobby contains one metacentric and 13 telocentric macrochromosomes and one metacentric and ten telocentric microchromosomes (2n = 50; FN = 54) and is similar to that described previously in 2n, but differs for macrochromosome morphology. Despite an about 40% difference in 2n and FN, these species have almost the same number of recombination nodules per genome: 51.4 ± 4.3 in the swift and 51.1 ± 6.7 in the hobby. The distribution of the recombination nodules along the macrochromosomes was extremely polarized in the Common Swift and was rather even in the Eurasian Hobby.
Conclusions
This study adds two more species to the short list of birds in which the number and distribution of recombination nodules have been examined. Our data confirm that recombination rate in birds is substantially higher than that in mammals, but shows rather a low interspecies variability. Even a substantial reduction in chromosome number does not lead to any substantial decrease in recombination rate. More data from different taxa are required to draw statistically supported conclusions about the evolution of recombination in birds.
Similar content being viewed by others
Background
Meiotic recombination is the main source of genetic variability in any population of sexually reproducing organisms. For this reason, the number of recombination events (crossovers) per genome (recombination rate) and their distribution along the chromosomes are considered as important genomic characteristics of species and studied actively in plants, fungi and animals (mostly mammals). These studies demonstrated substantial interspecies variation in recombination rate (Dumont and Payseur 2008; Frohlich et al. 2015; Dapper and Payseur 2017; Stapley et al. 2017) while within-population variation was found to be of the same magnitude across various taxa (Ritz et al. 2017). The minimum possible rate of recombination is constrained by the necessity of at least one crossover per pair of homologous chromosomes to ensure their orderly segregation in the first meiotic division. Thus, the recombination rate cannot be lower than the haploid chromosome number (n). Studies on mammals demonstrated that chromosome number (2n) and chromosome arm number (FN) are the best predictors of the recombination rate (Pardo-Manuel de Villena and de Sapienza 2001; Segura et al. 2013; Capilla et al. 2016). A well-known phenomenon of crossover interference (a low probability of crossovers to occur close to each other) is an important constraint of the maximum possible rate of recombination (Berchowitz and Copenhaver 2010; Segura et al. 2013). The smaller the chromosome, the less likely it contains more than one crossover. Thus, the total genome size and the total length of the synaptonemal complex (SC, the core structure of pachytene chromosomes) may also serve as predictors of species-specific recombination rate (Peterson et al. 1994; Kleckner et al. 2003). While molecular and cellular mechanisms controlling the recombination rate are more or less clear, the adaptive importance of interspecies differences for this trait remains a matter of discussion (Stapley et al. 2017; Dapper and Payseur 2017; Ritz et al. 2017).
Birds provide a good model to study the evolution of recombination. With rather a small genome size (about 1.4 pg) (Wright et al. 2014) and very conservative karyotypes (2n being about 78–82 and FN being 80–90 in most species) (Griffin and Burt 2014), they have undergone rapid speciation and evolved various adaptations to a wide variety of habitats. Unfortunately, only less than 10% of bird species have been karyotyped (Griffin and Burt 2014). Recombination traits have been studied in as few as nine species by cytological methods (see Table 1 and references therein) and in nine species by linkage analysis (Dawson et al. 2007; Groenen et al. 2009; Jaari et al. 2009; Aslam et al. 2010; Hansson et al. 2010; Kawakami et al. 2014; van Oers et al. 2014).
Most of the birds species studied for recombination rate have common karyotypes (2n = 78–82). Only two species with reduced chromosome number (2n = 68 and 74) have been examined and they did not show a drastic reduction of the recombination rate (Lisachov et al. 2017a). More birds with atypical karyotype as Apodidae (swifts), Psittaciformes (parrots) and Falconidae (falcons) need to be studied to reveal the cytogenetic and evolutionary basis of interspecies variation in recombination.
There are two reasons why bird meiotic chromosomes are poorly studied. First, the pachytene cells suitable for analysis can only be obtained in short time windows: from testes of adult males during or shortly before the breeding season or from ovaries of female embryos before hatching or chicks soon after hatching (Scanes 2014). Secondly, the bird has to be sacrificed.
In this study, we collected materials from fatally injured Common Swift (Apus apus Linnaeus, 1758) and Eurasian Hobby (Falco subbuteo Linnaeus, 1758) males provided by the local Bird of Prey Rehabilitation Centre. These specimens represent two bird families, which show a wide karyotypic variation. The karyotype of the Common Swift remains unknown. However, the other swifts examined have reduced and variable diploid numbers (from 62 to 70) (Yadav et al. 1995; Torres et al. 2004). Falconids show a very wide variation in diploid chromosome numbers (from 40 to 92) (Christidis 1990). Eurasian Hobby has 2n = 50 (Christidis 1990; Wang and Chen 1998; Nishida et al. 2008).
In this study, we for the first time describe the somatic and pachytene karyotypes of the male Common Swift and the pachytene karyotype of the male Eurasian Hobby. We estimate the overall number and distribution of recombination events along the chromosomes of these species, using immunolocalization of SYCP3, the protein of the SC lateral elements, MLH1, the mismatch repair protein marking mature recombination nodules, and centromere proteins. This method has been successfully used to analyze recombination landscapes in fish (Moens 2006; Lisachov et al. 2015), reptiles (Lisachov et al. 2017b, c), mammals (Capilla et al. 2016) and birds (Pigozzi 2016). This approach has lower resolution than linkage analysis at the fine scale level. However, cytological approach provides more reliable estimate of total number of global recombination rate, because it does not depend on the number and location of informative markers. Immunocytological analysis of SC spreads is especially efficient to examine bird karyotypes, because it provides unambiguous visualization of all microchromosomes, while at conventional and differential stained metaphase spreads it is difficult to distinguish microchromosomes from non-specifically stained debris.
Methods
Specimens
An adult male Common Swift with fatal accident trauma was provided by the Bird Rehabilitation Centre of Novosibirsk and euthanized in our laboratory. The testes of an adult Eurasian Hobby were collected immediately after the bird’s death in the Centre and transported to the laboratory in cold PBS. The birds were handled and euthanized in accordance with the approved national guidelines for the care and use of laboratory animals. All experiments were approved by the Ethics Committee on Animal Care and Use of the Institute of Cytology and Genetics of Siberian Department of the Russian Academy of Sciences, Russia (approval No. 35 of October 26, 2016). No additional permits are required for research on non-listed species in Russia.
Karyotyping
Mitotic chromosome preparations were obtained from short-term bone marrow cell culture of the Common Swift. The culture was incubated for 2 h at 37 °C in culture medium with 10 µg/mL colchicine. Hypotonic treatment was performed with 0.56% KCl solution for 15 min at 37 °C and followed by centrifugation for 5 min at 500×g. Fresh cold fixative solution (methanol: glacial acetic acid = 3:1 v/v) was changed tree times. Cell suspension was dropped on the wet cooled slides. The slides were dried for 2 h at 65 °C and stained for 4 min with 1 μg/mL solution of DAPI in 2× SSC. The slides were then washed in deionized water, dried at room temperature and mounted in Vectashield antifade mounting medium.
Spermatocyte spreading and immunostaining
Spermatocyte spreads were prepared from testes using a drying-down technique (Peters et al. 1997). Immunostaining was performed as described by Anderson et al. (1999). Primary antibodies used in this study were as follows: rabbit polyclonal to SYCP3 (1:500; ab150292, Abcam), mouse monoclonal to MLH1 (1:50; ab14206, Abcam), and human anti-centromere (ACA) (1:100, Cat #15-235-0001, Antibodies Inc.). The secondary antibodies used were Cy3-conjugated goat anti-rabbit (1:500; Cat#111-165-144, Jackson ImmunoResearch), FITC-conjugated goat anti-mouse (1:50; Cat#115-095-003, Jackson ImmunoResearch), and AMCA-conjugated donkey anti-human (1:100, Cat#709-155-098, Jackson ImmunoResearch). All antibodies were diluted in PBT (3% bovine serum albumin, 0.1% Tween 20 in phosphate buffered saline). A solution of 10% PBT was used to perform a blocking reaction. Primary antibody incubations were performed overnight in a humid chamber at 37°, secondary antibody incubations were performed for 1 h at 37 °C. Finally, slides were mounted in Vectashield with or without DAPI (Vector Laboratories).
The preparations were visualized with an Axioplan 2 imaging microscope (Carl Zeiss) equipped with a CCD camera (CV M300, JAI), CHROMA filter sets, and the ISIS4 image-processing package (MetaSystems GmbH). Brightness and contrast of all images were enhanced using Corel PaintShop Photo Pro X6 (Corel Corp).
Chromosome measurements and generation of recombination maps
The centromeres were identified by ACA foci. MLH1 signals were only scored if they were localized on SCs. The length of the SC of each chromosome arm was measured in micrometers and the positions of centromeres and MLH1 foci in relation to the centromere were recorded using MicroMeasure 3.3 (Reeves 2001). Individual SCs of macrochromosomes were identified by their relative lengths and centromeric indexes. To generate recombination maps of the macrochromosomes, we calculated the absolute position of each MLH1 focus multiplying the relative position of each focus by the average absolute length of the chromosome arm. These data were pooled for each arm and graphed to represent a recombination map. We measured absolute distances between two MLH1 foci within the arm in acrocentric and metacentric SCs and across the centromere in metacentric SCs. The relative distances were calculated as fractions of the SC length.
Statistica 6.0 software package (StatSoft) was used for descriptive statistics. All results were expressed as mean ± SD; p < 0.05 was considered as statistically significant.
Results
Karyotypes
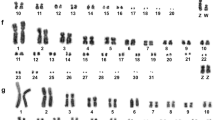
Somatic (Fig. 1a, b) and pachytene (Figs. 2a, 3a) karyotypes of the Common Swift consist of 39 chromosome pairs (2n = 78). Macrochromosomes 1, 3 and 5 are metacentrics, 2, 4 and 6 are submetacentrics, all the other chromosomes are telocentrics forming a row gradually decreasing in length.
SCs of the Common Swift (a) and Eurasian Hobby (b) after immunolocalization of SYCP3 (red), MLH1 (green) and centromere proteins (blue). The arrow points to metacentric microchromosome (m). The insert is a close-up of SC1. Arrowheads point to strong (s) and weak (w) centromere signals at the SC1. Bar: 10 µm
Pachytene karyotype of the Eurasian Hobby comprises 25 chromosome pairs (2n = 50). Its largest macrochromosome (SC1) and one of the microchromosomes are metacentrics, all the other chromosomes are telocentrics (Figs. 2b, 3b).
In the majority of pachytene cells (40 of 61), SC1 contained two distinct centromere signals 3.9 ± 1.6 µm away from each other. One was strong, similar to that of the other chromosome centromeres, the other was very weak. We have often observed MLH1 foci between the two centromere signals.
Apparently, one of the middle-sized macrochromosomes in both species is Z chromosome. However, it could not be identified for its size and morphology. In the Needle-tailed Swift (Hirundapus caudacutus), the species distantly related the Common Swift, Z chromosome is the smallest of metacentric macrochromosomes (Christidis 1990). In all Falco species examined it is middle-sized acrocentric chromosome (Nishida et al. 2008).
MLH1 focus number and distribution along macrochromosomes
To estimate the recombination rate and distribution of crossovers along the chromosomes of the species examined, we analyzed 1131 MLH1 foci at 858 SCs in 22 pachytene cells of the Common Swift and 3119 MLH1 foci at 1525 SCs in 61 pachytene cells of the Eurasian Hobby. The total length of SC in the swift was significantly smaller than that in the hobby (208.2 ± 31.8 and 257.9 ± 49.8 µm, correspondingly, p < 0.05).
The number of MLH1 foci per pachytene cell was almost the same in both species: 51.4 ± 4.3 in the swift and 51.1 ± 6.6 in the hobby, p > 0.05. To estimate the total length of the recombination map in centimorgans (cM), we multiplied the average number of MLH1 foci per cell by 50 map units (one recombination event = 50 cM). The resulting estimates were 2570 cM in the swift and 2555 cM in the hobby. Recombination rate (RR) measured as the ratio of the total genetic map length in cM to the genome size in Mb was also similar in both species. However, the recombination density (RD) estimated as the number of MLH1 foci per cell per 1 µm of SC was higher in the swift (0.25) than in the hobby (0.20) (Table 1).
The patterns of MLH1 distribution along the six largest macrochromosomes reveal drastic differences between swift and hobby (Fig. 4). The swift SCs demonstrated an extremely polarized distribution of MLH1 foci. Most of them are located at the distal chromosome ends (Fig. 4a). By contrast, the hobby SCs showed rather an even distribution of MLH1 foci. The frequency of the foci was somewhat elevated at both ends of the metacentric SC1 and acrocentric SC2-SC6. Polarization was more prominent in SC6, but still less pronounced than in the metacentric SC of the swift. Suppression of the MLH1 foci around the centromere, typical for most recombination landscapes described so far, was rather weak in the metacentric SCs of both species. In all acrocentric bivalents we observed an increased MLH1 focus frequency near the centromeres.
Distribution of MLH1 foci along individual SCs in pachytene spermatocytes of the Common Swift (a) and Eurasian Hobby (b). On the x-axis: the relative position of MLH1 foci at the six largest macroSCs in relation to the centromere (black triangle). The width of the interval is 1 μm. On the y-axis: the proportion of MLH1 focus number in each interval. Colors indicate bivalents with 1–9 MLH1 foci per bivalent. The scale shows the color codes
Crossover interference plays an important role in the determination of the number and distribution of crossovers along chromosomes (Berchowitz and Copenhaver 2010; Segura et al. 2013). In this study, we measured the interference by relative distances between the neighboring MLH1 foci at the macrochromosomes: the shorter the distance, the weaker the interference. Table 2 shows that the swift chromosomes accommodated less MLH1 foci than the hobby chromosomes of comparable size. The relative distances between the foci at the swift SCs were 1.5–2-fold longer than those at the hobby SCs. This is indicative of a higher crossover interference of the swift macrochromosomes.
Discussion
In this study, we for the first time describe the somatic and pachytene karyotypes of the male Common Swift and the pachytene karyotype of the Eurasian Hobby and estimate the overall number and distribution of recombination events along the chromosomes of these species.
The karyotype of the Common Swift consists of three metacentric, three submetacentric and two telocentric macrochromosomes and 31 telocentric microchromosomes (2n = 78; FN = 90). It differs from the karyotypes of the previously described related Apodidae species: Little Swift (Apus affinis affinis) (2n = ± 70) (Yadav et al. 1995), Fork-tailed Swift (Apus pacificus) (2n = 62), Needle-tailed Swift (H. caudacutus) (2n = 64) (Christidis 1990) and two tropical swift species in the genus Streptoprocne: S. biscutata (2n = 64) and S. zonaris (2n = 66) (Torres et al. 2004). Thus, the Common Swift has a karyotype that is typical of most Neoaves and apparently basal for Apodidae. The karyotype evolution of other swifts has probably involved fusions of some microchromosomes with each other or/and macrochromosomes and pericentric inversions of macrochromosomes (Torres et al. 2004). Apparently, the reduction of chromosome numbers occurred independently in the genera Apus, Hirundapus and Streptoprocne, which diverged from each other about 30–40 Mya (Jetz et al. 2012).
The karyotype of our specimen of the Eurasian Hobby coincides with those reported earlier for this species in the diploid number (2n = 50) but differs for the chromosome morphology (Christidis 1990; Nishida et al. 2008; Wang and Chen 1998). Its chromosome 1 and one of the microchromosomes are metacentrics, all the other chromosomes are telocentrics. Christidis (1990) referring to a conference report of X. Bian and Q. Li indicated that all chromosomes in the hobby karyotype were telocentric with exception of chromosome 1, which was acrocentric. Wang and Chen (1998) described the karyotypes of two hobby specimens from China. Both of them had 2n = 50, but differed in macrochromosome morphology. A male from Qiqihar city had metacentric chromosome 1, subtelocentric chromosomes 2, 4 and 5, submetacentric chromosome 9 and all the other chromosomes as telocentrics. A female from Dalian city differed from this specimen by the morphology of chromosomes 2, 4, and 5: they were telocentric. It is likely that this species is polymorphic for pericentric inversions or/and centromere shifts or/and the presence of additional heterochromatic short arms at some chromosomes.
It should be noted, however, that the somatic metaphase and pachytene SC karyotypes of the same specimen might differ in details or, to be precise, reveal different details of the chromosome structure. Immunolocalization of SYCP3 might misrepresent the arm ratio of some chromosomes. It is well known that heterochromatin is underrepresented in the SC (Stack 1984). Studies on mammals show that short heterochromatic arms present at some chromosomes may not be visualized at SCs (Graphodatsky et al. 2000). On the other hand, immunolocalization of centromere proteins reveals centromere position more reliably than primary constrictions at metaphase spreads. For example, the metacentric microchromosome detected in our study and absent in the description of Wang and Chen (1998) can only be identified at pachytene spreads (Fig. 2b), but not at metaphase plates.
Two centromere signals that we detected at the hobby SC1 might indicate heterozygosity for pericentric inversion or a centromere shift, or the presence of vestigial inactive centromere sequence at one or both homologs. Although pericentric inversions and centromere shifts are well documented mechanisms of bird chromosome evolution (Kasai et al. 2003; Skinner and Griffin 2012; Griffin and Burt 2014), it is unlikely that they are the cause of the double centromere signal in the hobby. The occurrence of MLH1 foci between two centromere signals disapproves inversion heterozygosity, and the difference in signal intensity disapproves centromere shift. If there had been inversion heterozygosity, no crossovers would have occurred in the non-homologously synapsed inverted region. If there had been centromere shift, both signals would have been of the same intensity and weaker than the signals of the centromeres of other chromosomes. The vestigial inactive centromere sequence is the most likely explanation of the observed feature. Inactive centromeres in yeasts, plants and mammals lack several centromere proteins, such as CENP-A, CENP-C and CENP-E (Ross et al. 2015). If that were the case with the bird chromosomes, that would explain the weaker signal of the secondary centromere at the hobby SC1.
A rather surprising result of our study is that despite an about 40% difference in 2n and FN, the swift and the hobby have almost the same number of recombination nodules per genome. Apparently, the decrease of the lower limit of recombination rate (the number of obligatory crossovers) in the hobby has been compensated by a relaxation of the higher limit: increase in SC length and decrease in crossover interference. Similar in overall recombination rate as they are, the swift and hobby have very different patterns of recombination event distribution along macrochromosomes: extremely polarized in the swift and rather even in the hobby. A relatively even crossover distribution along the macrochromosomes with moderate peaks at the chromosome ends (including pericentromeric ends of acrocentrics) has been observed in the majority of bird species examined cytologically. A polarized distribution is more typical of mammalian chromosomes (Borodin et al. 2009; Segura et al. 2013) and macrochromosomes of anole lizards (Lisachov et al. 2017c). In birds, a pattern like this has so far been described in Zebra Finch and is considered as unique (Calderon and Pigozzi 2006; Backström et al. 2010).
Conclusions
Our data add two more species to the list of birds in which the number and distribution of recombination nodules have been examined (Table 1). This list is short; however, it already reveals some important features of recombination in birds. It confirms earlier suggestions that recombination rate in birds is substantially higher than in mammals. At the same time, birds demonstrate rather a low interspecies variably of recombination rate compared to mammals. This may in part be due to a strong conservation of chromosome number in birds. However, our data demonstrate that reduction of chromosome number does not lead to decrease of recombination rate. Meanwhile, more data from different taxa are needed to draw statistically supported conclusions about the evolution of recombination in birds.
References
Anderson LK, Reeves A, Webb LM, Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 1999;151:1569–79.
Aslam ML, Bastiaansen JWM, Crooijmans RPMA, Vereijken A, Megens H-J, Groenen MAM. A SNP based linkage map of the turkey genome reveals multiple intrachromosomal rearrangements between the turkey and chicken genomes. BMC Genom. 2010;11:647.
Backström N, Forstmeier W, Schielzeth H, Mellenius H, Nam K, Bolund E, Webster MT, Öst T, Schneider M, Kempenaers B, Ellegren H. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 2010;20(4):485–95.
Berchowitz LE, Copenhaver GP. Genetic interference: don’t stand so close to me. Curr Genom. 2010;11:91–102.
Borodin PM, Basheva EA, Zhelezova AI. Immunocytological analysis of meiotic recombination in the American mink (Mustela vison). Anim Genet. 2009;40:235–8.
Calderon PL, Pigozzi MI. MLH1-focus mapping in birds shows equal recombination between sexes and diversity of crossover patterns. Chromosome Res. 2006;14:605–12.
Capilla L, Garcia Caldés M, Ruiz-Herrera A. Mammalian meiotic recombination: a toolbox for genome evolution. Cytogenet Genome Res. 2016;150:1–6.
Christidis L. Animal cytogenetics 4: Chordata 3; B, Aves. Berlin: Gebrüder Borntraeger; 1990.
Dapper AL, Payseur BA. Connecting theory and data to understand recombination rate evolution. Philos Trans R Soc B Biol Sci. 2017. https://doi.org/10.1098/rstb.2016.0469.
Dawson DA, Åkesson M, Burke T, Pemberton JM, Slate J, Hansson B. Gene order and recombination rate in homologous chromosome regions of the chicken and a passerine bird. Mol Biol Evol. 2007;24:1537–52.
del Priore L, Pigozzi MI. Broad-scale recombination pattern in the primitive bird Rhea americana (Ratites, Palaeognathae). PLoS ONE. 2017;12:e0187549.
Dumont BL, Payseur BA. Evolution of the genomic rate of recombination in mammals. Evolution. 2008;62:276–94.
Frohlich J, Vozdova M, Kubickova S, Cernohorska H, Sebestova H, Rubes J. Variation of meiotic recombination rates and MLH1 foci distribution in spermatocytes of cattle, sheep and goats. Cytogenet Genome Res. 2015;146:211–21.
Graphodatsky AS, Sablina OV, Meyer MN, Malikov VG, Isakova EA, Trifonov VA, Polyakov AV, Lushnikova TP, Vorobieva NV, Serdyukova NA, Perelman PL, Borodin PM, Benda P, Frynta D, Leikepová L, Munclinger P, Piálek J, Sádlová J, Zima J. Comparative cytogenetics of hamsters of the genus Calomyscus. Cytogenet Genome Res. 2000;88:296–304.
Gregory TR. Animal genome size database. 2017. http://www.genomesize.com. Accessed 9 Nov 2017.
Griffin D, Burt DW. All chromosomes great and small: 10 years on. Chromosome Res. 2014;22:1–6.
Groenen MAM, Wahlberg P, Foglio M, Cheng HH, Megens H-J, Crooijmans RPMA, Besnier F, Lathrop M, Muir WM, Wong GKS, Gut I, Andersson L. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res. 2009;19:510–9.
Hansson B, Ljungqvist M, Dawson DA, Mueller JC, Olano-Marin J, Ellegren H, Nilsson J-Å. Avian genome evolution: insights from a linkage map of the blue tit (Cyanistes caeruleus). Heredity. 2010;104:67–78.
Jaari S, Li M-H, Merilä J. A first-generation microsatellite-based genetic linkage map of the Siberian jay (Perisoreus infaustus): insights into avian genome evolution. BMC Genom. 2009;10:1.
Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012;491:444–8.
Kasai F, Garcia C, Arruga MV, Ferguson-Smith MA. Chromosome homology between chicken (Gallus gallus domesticus) and the red-legged partridge (Alectoris rufa); evidence of the occurrence of a neocentromere during evolution. Cytogenet Genome Res. 2003;102:326–30.
Kawakami T, Smeds L, Backström N, Husby A, Qvarnström A, Mugal CF, Olason P, Ellegren H. A high-density linkage map enables a second-generation collared flycatcher genome assembly and reveals the patterns of avian recombination rate variation and chromosomal evolution. Mol Ecol. 2014;23:4035–58.
Kleckner N, Storlazzi A, Zickler D. Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet. 2003;19:623–8.
Lisachov AP, Malinovskaya LP, Druzyaka AV, Borodin PM, Torgasheva AA. Synapsis and recombination of autosomes and sex chromosomes in two terns (Sternidae, Charadriiformes, Aves). Vavilovskii Zhurnal Genet Selektsii. 2017a;21:259–68 (in Russian).
Lisachov AP, Trifonov VA, Giovannotti M, Ferguson-Smith MA, Borodin PM. Heteromorphism of “homomorphic” sex chromosomes in two anole species (Squamata, Dactyloidae) revealed by synaptonemal complex analysis. Cytogenet Genome Res. 2017b;151:89–95.
Lisachov AP, Trifonov VA, Giovannotti M, Ferguson-Smith MA, Borodin PM. Immunocytological analysis of meiotic recombination in two anole lizards (Squamata, Dactyloidae). Comp Cytogenet. 2017c;11:129–41.
Lisachov AP, Zadesenets KS, Rubtsov NB, Borodin PM. Sex chromosome synapsis and recombination in male guppies. Zebrafish, 2015,12:174–80.
Moens PB. Zebrafish: chiasmata and interference. Genome. 2006;49:205–8.
Nishida C, Ishijima J, Kosaka A, Tanabe H, Habermann FA, Griffin DK, Matsuda Y. Characterization of chromosome structures of Falconinae (Falconidae, Falconiformes, Aves) by chromosome painting and delineation of chromosome rearrangements during their differentiation. Chromosome Res. 2008;16:171–81.
Pardo-Manuel de Villena F, de Sapienza C. Recombination is proportional to the number of chromosome arms in mammals. Mamm Genome. 2001;12:318–22.
Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–8.
Peterson DG, Stack SM, Healy JL, Donohoe BS, Anderson LK. The relationship between synaptonemal complex length and genome size in four vertebrate classes (Osteicthyes, Reptilia, Aves, Mammalia). Chromosome Res. 1994;2:153–62.
Pigozzi MI. Distribution of MLH1 foci on the synaptonemal complexes of chicken oocytes. Cytogenet Cell Genet. 2001;95:129–33.
Pigozzi MI. The chromosomes of birds during meiosis. Cytogenet Genome Res. 2016;150:128–38.
Pigozzi MI, del Priore L. Meiotic recombination analysis in female ducks (Anas platyrhynchos). Genetica. 2016;144:307–12.
Pigozzi MI, Solari AJ. Equal frequencies of recombination nodules in both sexes of the pigeon suggest a basic difference with eutherian mammals. Genome. 1999;42:315–21.
Reeves A. MicroMeasure: a new computer program for the collection and analysis of cytogenetic data. Genome. 2001;44:439–43.
Ritz KR, Noor MAF, Singh ND. Variation in recombination rate: adaptive or not? Trends Genet. 2017;33:364–74.
Ross J, McNulty S, Sullivan B. The epigenetics of centromere function. In: Chadwick BP, editor. Epigenetics. Norfolk: Caister Academic Press; 2015. p. 133–66.
Scanes CG. Sturkie’s avian physiology. 6th ed. London: Academic Press; 2014.
Segura J, Ferretti L, Ramos-Onsins S, Capilla L, FarréM Reis F, Oliver-Bonet M, Fernández-Bellón H, Garcia F, Garcia-Caldés M, Robinson TJ, Ruiz-Herrera A. Evolution of recombination in eutherian mammals: insights into mechanisms that affect recombination rates and crossover interference. Proc R Soc B Biol Sci. 2013;280:1945.
Skinner BM, Griffin DK. Intrachromosomal rearrangements in avian genome evolution: evidence for regions prone to breakpoints. Heredity. 2012;108:37–41.
Stack SM. Heterochromatin, the synaptonemal complex and crossing over. J Cell Sci. 1984;71:159–76.
Stapley J, Ferretti L, Ramos-Onsins S, Capilla L, Farré M, Reis F, Oliver-Bonet M, Fernández-Bellón H, Garcia F, Garcia-Caldés M, Robinson TJ, Ruiz-Herrera A. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Philos Trans R Soc Lond B. 2017;372:20160455.
Torgasheva AA, Borodin PM. Immunocytological analysis of meiotic recombination in the gray goose (Anser anser). Cytogenet Genome Res. 2017;151:27–35.
Torres RA, Ribeiro J, Adam ML. Genetic discontinuity and evolutionary scenario among Neotropical and Old World swift species (Aves: Apodiformes: Apodidae). Caryologia. 2004;57:177–83.
van Oers K, Santure AW, De Cauwer I, van Bers NEM, Crooijmans RPMA, Sheldon BC, Visser ME, Slate J, Groenen MAM. Replicated high-density genetic maps of two great tit populations reveal fine-scale genomic departures from sex-equal recombination rates. Heredity. 2014;112:307–16.
Wang ML, Chen FY. Study on karyotype of Hobby (Falco subbuteo) and Kestrel (Falco tinnunculus). J Forest Res. 1998;9:108–10.
Wright NA, Gregory TR, Witt CC. Metabolic “engines” of flight drive genome size reduction in birds. Proc R Soc B. 2014. https://doi.org/10.1098/rspb.2013.2780.
Yadav JS, Arora RB, Yadav AS. Karyotypes of two species of Indian birds and localization of nucleolus organizer regions. Cytobios. 1995;82:159–69.
Authors’ contributions
AT and PB provided the research idea and designed the experiments. LM, ES, and AT conducted the experiments and collected the data. LM and AT finished the data analysis and compiled the results. AT and PB supervised the research and wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We thank the Microscopic Center of the Siberian Department of the Russian Academy of Sciences for granting access to microscopic equipment.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval
The birds were handled and euthanized in accordance with the approved national guidelines for the care and use of laboratory animals. All experiments were approved by the Ethics Committee on Animal Care and Use of the Institute of Cytology and Genetics of Siberian Department of the Russian Academy of Sciences, Russia (approval No. 35 of October 26, 2016). No additional permits are required for research on non-listed species in Russia.
Funding
This work was supported by the Russian Foundation for Basic Research (Grant # 16-04-00087) and the Federal Agency for Scientific Organizations via the Institute of Cytology and Genetics (Grant # 0324-2018-0019). The funding bodies play no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Malinovskaya, L., Shnaider, E., Borodin, P. et al. Karyotypes and recombination patterns of the Common Swift (Apus apus Linnaeus, 1758) and Eurasian Hobby (Falco subbuteo Linnaeus, 1758). Avian Res 9, 4 (2018). https://doi.org/10.1186/s40657-018-0096-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40657-018-0096-7