Abstract
Acute coronary syndrome (ACS) is a clinical condition caused by a disturbance in myocardial blood flow. ACS can be basically divided into two forms: ST elevation myocardial infarction (STEMI) due to complete occlusion of the coronary artery and non-ST elevation myocardial infarction (NSTEMI) due to partial occlusion of the coronary artery. In this study, we aimed to monitor the metabolite profile of STEMI and NSTEMI patients and compare the results via untargeted metabolomics approach. Serum samples were collected from STEMI and NSTEMI patients, and each group consists of 20 participants. Extraction was achieved by acetonitrile, and chromatographic separation was performed by LC/Q-TOF/MS/MS accompanied with dual AJS ESI positive ion mode. METLIN, MATLAB 2017a-PLS Toolbox7.2, and Human Metabolome Database were utilized for bioinformatics evaluation of obtained findings. In our results, 203 m/z ratio was detected and 163 m/z ratio passed the significance criteria (fold analysis > 1.5 and p < 0.05). Twenty-five metabolites including BCAAs, LysoPC species, lactic acid, succinate, malonic acid, maleic acid, butyric acid, carnitine, and betaine were identified. In conclusion, new biomarker candidates were identified to differentiate the diagnosis of STEMI and NSTEMI. Identified metabolites are indicative of alterations in oxidative stress, hypoxia, TCA cycle, and amino acid metabolism.
Similar content being viewed by others
Introduction
Acute coronary syndromes (ACSs) are caused by erosion or sudden rupture of atherosclerotic plaque in the coronary artery wall (National CGCU 2013) and are still the most common name of death and disability of cardiovascular disease today (Damhoff and Huecker 2018). ACSs can be divided into subgroups of ST segment elevation myocardial infarction (STEMI), non-ST segment elevation myocardial infarction (NSTEMI), and unstable angina (UA) (Damhoff and Huecker 2018). The diagnosis of ACSs is based on the clinical history, electrocardiogram (ECG), and serum enzymes and troponin levels. Cardiac troponin is a protein released by infarcting myocardial cells and evaluated as critical markers in acute myocardial infarction with a concentration above the 99th percentile from reference value of cardiac troponin within the first 9 h according to the Joint European Society of Cardiology-American College of Cardiology Committee for the Redefinition of Myocardial Infarction. In some cases, patients who exhibit symptoms consistent with ACSs whereas normal or undetermined ECG and negative troponin could be observed that is commonly diagnosed as unstable angina (National CGCU 2013). Symptoms of NSTEMI show the similar features of unstable angina. So, diagnosis of NSTEMI can be typically made with the positive cardiac biomarkers in serum. STEMI diagnoses with the rise line are called as “ST-segment” on an ECG. There is no evidence of ST segment elevation in NSTEMI. So, it can be said that STEMI can be only separated from NSTEMI with ECG changes because the troponin increases are in both of them (Agewall et al. 2010; Laborde et al. 2014). There are no serum biomarkers to differentiate STEMI from NSTEMI. So, these shortcomings for predicting as STEMI or NSTEMI need clinically valuable new biomarkers to clarify the mechanisms of myocardial infarction.
Metabolomics can be described as the determination of all low molecular weight metabolites present in human biochemistry change depending on the physiological state (Yin et al. 2017). Nowadays, metabolomics studies are applied by using recently developed different analytical methods (LC-MS, GC-MS, and NMR) for the search for biomarkers in different biological liquids (urine, plasma, serum, saliva, and biopsy samples) of healthy and/or patients in human studies. In this context, it can be useful to differentiate STEMI from NSTEMI by enabling high-throughput profiling of large numbers of metabolites in serum.
There are several untargeted metabolomics studies for observing the metabolic alterations with carrying out by advanced spectroscopic methods such as GC-MS (Laborde et al. 2014; Ali et al. 2016; Yao et al. 2010), NMR (Ali et al. 2016; Bodi et al. 2012), LC/MS/MS (Huang et al. 2016), LC/Q-TOF/MS/MS (Zhu et al. 2018; Senn et al. 2012; Surendran et al. 2018), and CE/MS (Naz et al. 2015) in both STEMI (Laborde et al. 2014; Yao et al. 2010; Bodi et al. 2012; Huang et al. 2016; Zhu et al. 2018; Senn et al. 2012; Surendran et al. 2018; Naz et al. 2015; Lu et al. 2017; Xu et al. 2016) and NSTEMI (Laborde et al. 2014; Naz et al. 2015; Calderón-Santiago et al. 2014) separately. Among these studies, there are studies on the comparison of serum metabolite profiling between the STEMI and unstable angina (Ali et al. 2016; Lu et al. 2017; Xu et al. 2016) or between the stable angina and NSTEMI (Calderón-Santiago et al. 2014) with untargeted metabolomics approach with GC/MS (Ali et al. 2016; Lu et al. 2017) and LC-Q-TOF MS (Xu et al. 2016; Calderón-Santiago et al. 2014) which are available. In addition to this, only one study demonstrated the serum metabolite profiling of STEMI and NSTEMI with CE-MS (Naz et al. 2015) which is especially suitable for the determination of highly polar or ionic compounds. However, there is no study to compare the metabolic alterations between STEMI and NSTEMI via LC/Q-TOF/MS/MS which provides specific and comprehensive analysis of metabolites for both low and high hydrophobic molecules (Senn et al. 2012; Ussher et al. 2016).
The aim of this work is to investigate the serum samples of STEMI and NSTEMI patients via LC/Q-TOF/MS/MS method. Thus, crucial information could be obtained for both diseases. By the help of multivariate data analysis, any alteration between both cases could be determined and potential biomarkers identified for diagnosis.
Materials and methods
Data collection
Ethical committee approval was obtained from the medical faculty of Atatürk University Erzurum, Turkey, on 25 April 2018 with the number of 163, and written informed consent was received from all patients.
Twenty patients for each group who suffered from STEMI or NSTEMI were accepted into the study (the patients who have been diagnosed with myocardial infarction (MI) in the last 6 months). Exclusion criteria for patients were as follows: severe valve disease, chronic heart failure, coronary artery disease, atrial fibrillation, coronary bypass, malignancy, and chronic renal failure whose age ranged between 45 and 60. Patients were followed by the Department of Cardiology Clinic of Atatürk University. Both groups were evaluated for active smoking, diabetes mellitus, hypertension, and hyperlipidemia. Both groups were followed by the same cardiologist, and serum samples were taken from all patients. Routine hemogram and biochemistry data were calculated as samples were kept in deep freezer until chromatographic analysis.
Preparation of plasma samples
Serum samples were gently thawed on ice in order to avoid metabolic reactions and homogenized on vortex mixer. One hundred microliters of serum samples was spiked into Eppendorf vials, and protein precipitation was performed via 300 μL of acetonitrile. Samples were vigorously mixed about 1 min and kept on ice for 15 min in order to smash cell membranes and make metabolites free. Centrifugation was carried out at 16000g for 10 min. Approximately, 250 μL supernatants were taken and transferred to Eppendorf vials in order to evaporate the organic phase until dryness. Precipitates were re-dissolved in a mixture of mobile phase including acetonitrile and 0.1% formic acid containing 30 ng/mL of internal standard (IS), carbemazapine. IS was employed to increase the quality of data and to test the autonomous integration success of the METLIN. In addition to this, quality control (QC) samples were prepared by taking 5 μL of each extract. Blank sample was also analyzed to detect any interferences and false peaks. QC samples were injected after every 8 samples along the sequence for evaluating the repeatability of proposed analysis.
LC/Q-TOF/MS/MS parameters
An Agilent 6530 Accurate-Mass Q-TOF mass spectrometer (Agilent, USA) coupled with Agilent 1290 Infinity LC system and B.02.00 Agilent Mass Hunter Software (Agilent, USA) was used to implement this study. Dual Agilent Jet Stream Electrospray Ionization (AJS ESI) ion mode was selected for the Q-TOF mass spectrometer system, and the system was initiated in positive capillary voltage of 3.5 kV in positive ion mode; drying gas flow was 12 L/min, while gas temperature was set at 400 °C. The nebulizer pressure was arranged as 50 psig. The fragmentor voltage of the proposed method was set at 120 V. Accurate mass measurements during the analysis were obtained through an automated calibrate delivery system using a jet stream thermal focusing ESI source providing a low flow (100 mL/min) of a calibrating solution. Hypersil C18 (100 mm × 2.1 mm, 1.9 μm) column and graduate elution were chosen for chromatographic separation. Analysis time was arranged to be 15 min.
Bioinformatic and statistical analysis
Blank, QC, and blank plasma sample chromatograms were analyzed via Agilent Software, and then for peak alignment and correction, obtained data were transferred to XCMS Software. Following criteria were evaluated for each m/z ratio as p ≤ 0.01 and fold analysis > 1.5, and values that pass these parameters were taken into account to find potential biomarker metabolites for separating STEMI and NSTEMI cases. Identification of them was performed by comparing the obtained results with Human Metabolome Database (HMDb) and METLIN database. MS/MS spectra of each score were also evaluated to confirm identified metabolites. Classification of extracted metabolites was achieved via unsupervised OPLS-DA algorithm in MATLAB R2017a. Metabolic pathway analysis was performed by using MetaboAnalyst 4.0. Demographic features and laboratory parameters from STEMI and NSTEMI patients were also statistically compared with independent sample Student’s t test and χ2 or Fisher’s exact test by using the SPSS 24 software. p value lower than 0.05 was accepted as statistically significant.
Results
Characteristics of volunteers
Demographic features such as age, gender, and BMI and laboratory parameters such as CRP, HDL, LDL, TG, Troponin, and WBC values of STEMI and NSTEMI patients are presented in Table 1. According to the data obtained, no significant differences between demographic features of each group (p > 0.05) were determined whereas WBC and hypertension values were statistically different (p < 0.05).
Metabolite identification
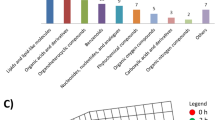
Two criteria were mostly used to identify metabolites which are mass to charge ratio and retention time. All blank, QC, and plasma sample chromatograms were exported to Agilent Mass Hunter Software. Obtained data were uploaded into XCMS software program. In this study, 203 m/z ratio were determined by the detector. Significance was determined as the fold analysis > 1.5 and p ≤ 0.01 criteria from m/z scores. Values that have passed these criteria were deeply investigated for metabolite identification. It is measured that 163 m/z signal provides these conditions (Fig. 1). Unfortunately, many of the m/z scores were encountered in more than one metabolite. Debates in m/z score were figured out by checking MS/MS results of blank and QC samples for eliminating false peaks and MS/MS spectra of each one. Twenty-five different metabolites were identified, these metabolites were monitored in Table 2, and matched metabolites were underlined via up and down symbols. So, MetaboAnalyst 4.0 was preferred for the detailed pathway analysis regarding the identified 25 metabolites. Determined pathways and their impacts are exhibited in Fig. 2. According to the MetaboAnalyst pathway analysis software, identified metabolites may have influence on 42 different pathways. The following numbers of metabolites were matched with related pathways: 7/75 aminoacyl t-RNA biosynthesis; 3/27 valine, leucine, and isoleucine biosynthesis; 2/20 TCA cycle; 2/32 glycerophospholipid metabolism; 2/ 24 alanine, aspartate, and glutamate metabolism; 3/40 butanoate metabolism; 3/39 nitrogene metabolism; 4/35 propanoate metabolism; 1/32 piruvate metabolism; and 3/48 glycine, serine, and threonine metabolism.
Chemometric analysis for metabolomics data
The main goal of metabolomics studies is to monitor a global snapshot of metabolites without observational bias in any biological medium or other analytical situations. Though such high information content is encountered in a challenge extracting as much as significant biological data, possible conclusions from any dataset indeed require advanced forms of data analyses matrix form into (Worley and Powers 2013). In order to maintain this data reduction in a trustworthy way, different chemometric approaches and data analyses were reported. One of the most preferred algorithms was OPLS-DA where classes are labeled in Y as a dummy variable and m/z data were added into X block. These two blocks utilized the metabolomics study matrix and cluster analysis which were performed via the mentioned datasets.
Predictivity of model was controlled via cross-validation assessment. Venetian blinds with 10 splits and 1 sample per split were applied to calculate cross-validation. Two latent variables exhibit two clusters. RMSEC values were calculated as 0.05 while square root of regression was 0.99. RMSEC confirms the goodness of fit for the method. LV1 vs LV2 graph are plotted and monitored in Fig. 3. Hierarchical cluster analyses were also plotted in order to exhibit the inter-relation of each sample. Ward’s method was applied to TOF MS/MS data, and two clusters were successfully obtained. Dendogram is monitored in Fig. 4. According to the chemometric models, successful diagnosis rate could be obtained by just analyzing the plasma samples via TOF MS/MS to make decision about either STEMI or NSTEMI.
Discussion
MS is an analytical method of focusing on mass to charge ratio (m/z) and abundance of ions at high speed of vacuum conditions. It is generally accepted that MS is one of the most powerful probes into the identification of matter. MS could be used with both gas (Laborde et al. 2014; Ali et al. 2016; Yao et al. 2010) and liquid chromatography (Huang et al. 2016; Zhu et al. 2018; Surendran et al. 2018; Naz et al. 2015) and capillary electrophoresis (Naz et al. 2015). In this study, liquid chromatography was preferred to get as much data as possible. Because gas chromatography has some limitations, only volatile compounds could be analyzed. Furthermore, an LC/Q-TOF/MS/MS instrumentation was chosen to gain a widespread profiling for the identification of endogenous metabolites. By the help of multivariate data analysis, metabolic differences between STEMI and NSTEMI patients were evaluated.
In the present study, almost 203 m/z ratio was determined by the detector. Features that provide the fold analysis > 1.5 and p ≤ 0.01 criteria from m/z scores were considered to be new biomarker candidate metabolites. One hundred sixty-three m/z signal passed this criteria (Fig. 1). Among them, 25 potential metabolites and their related metabolic pathways were taken into account (Table 2 and Fig. 2).
The major recognized several metabolites and, of course, pathways including energy metabolism, amino acid metabolism, fatty acid oxidation, glycerophospholipid synthesis, anaerobic glycolysis, and urea cycle were differentiated, and most of them were related with ischemia and oxidative stress.
There is a strong correlation between branched-chain amino acids (BCAAs) and cardiovascular AEs after STEMI may be due to the abnormal BCAA catabolism and MI and ischemia–reperfusion injury (Naz et al. 2015; Du et al. 2018). BCAAs, such as leucine, isoleucine, lysine, alanine, and l-valine, are known as essential amino acids supplied by diet (Harper et al. 1984). Despite elevated dietary protein intake may contribute to high circulating levels of BCAAs, recent studies have shown that higher plasma BCAA concentrations in individuals with cardiovascular disease are brought about by obstructed BCAA catabolism in the myocardium, which results in a build-up of myocardial BCAA that spills over into the circulation (Wang et al. 2011). In our untargeted metabolomics study, it is measured that l-valine, leucine, isoleuicine, lysine, and alanine were significantly high in STEMI patients with respect to the NSTEMI. It could be claimed that STEMI and NSTEMI mechanisms are different and high BCAA concentration may cause STEMI. So people who have cardiovascular diseases may change their diet avoiding excess amount of BCAA-rich nutrition.
The creatine kinase system protects the cardiovascular system from ischemia and increases contractility. So, increased creatine indicates a decrease in the activity of the creatine kinase system (Bottomley et al. 2013). In this study, we found that creatine was lower in NSTEMI compared to STEMI.
Carnitine level and STEMI/NSTEMI diseases were also related. The elevated concentration of carnitine in the blood plasma of STEMI/NSTEMI patients might have resulted due to the insufficient uptake and/or increased leakage of carnitine along the ischemic myocardium (Khan et al. 2014). Carnitine was also found lower in NSTEMI compared to STEMI.
In previous studies, LysoPC has been positively associated with inflammation and atherosclerosis and LysoPC species were also evaluated as potential biomarkers for STEMI/NSTEMI (Pouralijan Amiri et al. 2019). In our experiments, it was found that LysoPC (18:0) was lower in STEMI, while LysoPC (18:1) level was high in NSTEMI.
Oxidative stress is known to play an important role in various inflammatory diseases and atherosclerosis (Dhalla et al. 2000). Oxidative stress plays an important role in ischemic myocardial injury, and elevated levels of ROS have been detected in ischemic myocardium (Misra et al. 2009). Several studies have reported that butyric acid, maleic acid, and malonic acid cause oxidative stress and fumaric acid was an endogenous antioxidant compound (Laborde et al. 2014; Maragos et al. 2004; Wu et al. 2017). Thus, increased butyric acid, maleic acid, and malonic acid and decreased fumaric acid can have an important role in ischemic myocardial injury and may be related to the degree of ischemia. In this study, we found that butyric acid, maleic acid, and malonic acid were increased in STEMI compared to NSTEMI.
Previous studies suggest that the role of citric acid cycle intermediates such as lactate, glucose, aminobutyric acid, citrulline, and argino-succinateas are key metabolic mediators of acute myocardial ischemia (Barba et al. 2008; Goonewardena et al. 2010). In this study, similarly, we found that lactate, succinate, and malonate were lower in NSTEMI versus STEMI.
Our study also showed that several acids such as lactic acid, malonic acid, maleic acid, carbamic acid, and benzoic acid and hydroxylamine levels were significantly higher in STEMI patients against NSTEMI group. These molecules play an important role in nitrogen metabolism, and highness in their concentration could be related to oxidative stress (Rustom et al. 2003). More hypoxic and acidotic situation, that is an expected case in, could make an increase on some acidic metabolites (Graham et al. 2004). STEMI acidosis levels were certainly high in accordance with proposed findings.
Betaine level was also associated with cardiovascular diseases, and this metabolite was found much lower in STEMI patients.
It is obvious that this study contains some restrictions. Sample size could be wider regarding to highly prevalent disease, and we could not obtained serum samples of patient group after percutaneous coronary intervention (PCI). Secondly, metabolite concentration may be affected by diet, drug consumption, smoking, and presence of other chronic diseases. Lastly, metabolomics approach deals with discovering new biomarkers or illuminating a pathway that is crucial for understanding the mechanism of any disorder. Thus, such methods generally focus on qualitative analysis. Determining the exact concentration of metabolites is impossible in such processes. Semi-quantitative results give only differences between groups. Exact information for the concentration of any metabolites could not be obtained by this method.
Conclusions
The proposed untargeted metabolomics study is the first TQ-TOF-MS/MS-based work for comparing the metabolic alterations of STEMI and NSTEMI patients. Differentiations of each group were succeeded in via OPLS-DA. According to data obtained, there is a statistically significant difference between BCAAs, LysoPC species, lactic acid, succinate, malonic acid, maleic acid, butiric acid, carnitine, and betaine levels for each group. Study showed that excess dietary intake of BCAA may cause high risk on STEMI. In addition to this, diagnosis of STEMI could be achieved by evaluating the metabolic profile of patients.
Availability of data and materials
Not applicable
Abbreviations
- ACS:
-
Acute coronary syndrome
- BCAAs:
-
Branched-chain amino acids
- ECG:
-
Electrocardiogram
- IS:
-
Internal standard
- MI:
-
Myocardial infarction
- NSTEMI:
-
Non-ST elevation myocardial infarction
- PCI:
-
Percutaneous coronary intervention
- QC:
-
Quality control
- STEMI:
-
ST elevation myocardial infarction
- UA:
-
Unstable angina
References
Agewall S, Giannitsis E, Jernberg T, Katus H. Troponin elevation in coronary vs. non-coronary disease. Eur Heart J. 2010;32(4):404–11.
Ali SE, Farag MA, Holvoet P, Hanafi RS, Gad MZ. A comparative metabolomics approach reveals early biomarkers for metabolic response to acute myocardial infarction. Sci Rep. 2016;6:36359.
Barba I, de Leon G, Martín E, Cuevas A, Aguade S, Candell-Riera J, Barrabés JA, Garcia-Dorado D. Nuclear magnetic resonance-based metabolomics predicts exercise-induced ischemia in patients with suspected coronary artery disease. Magn Reson Med. 2008;60(1):27–32.
Bodi V, Sanchis J, Morales JM, Marrachelli VG, Nunez J, Forteza MJ, Chaustre F, Gomez C, Mainar L, Minana G. Metabolomic profile of human myocardial ischemia by nuclear magnetic resonance spectroscopy of peripheral blood serum: a translational study based on transient coronary occlusion models. J Am Coll Cardiol. 2012;59(18):1629–41.
Bottomley PA, Panjrath GS, Lai S, Hirsch GA, Wu K, Najjar SS, Steinberg A, Gerstenblith G, Weiss RG. Metabolic rates of ATP transfer through creatine kinase (CK Flux) predict clinical heart failure events and death. Sci Transl Med. 2013;5(215):215re213.
Calderón-Santiago M, Priego-Capote F, Galache-Osuna J, de Castro ML. Analysis of serum phospholipid profiles by liquid chromatography–tandem mass spectrometry in high resolution mode for evaluation of atherosclerotic patients. J Chromatogr A. 2014;1371:154–62.
Damhoff TC, Huecker MR. Myocardial infarction, non ST segment elevation (NSTEMI). StatPearls: StatPearls publishing; 2018.
Dhalla NS, Temsah RM, Netticadan T. Role of oxidative stress in cardiovascular diseases. J Hypertens. 2000;18(6):655–73.
Du X, You H, Li Y, Wang Y, Hui P, Qiao B, Lu J, Zhang W, Zhou S, Zheng Y. Relationships between circulating branched chain amino acid concentrations and risk of adverse cardiovascular events in patients with STEMI treated with PCI. Sci Rep. 2018;8(1):15809.
Goonewardena SN, Prevette LE, Desai AA. Metabolomics and atherosclerosis. Curr Atheroscler Rep. 2010;12(4):267–72.
Graham RM, Frazier DP, Thompson JW, Haliko S, Li H, Wasserlauf BJ, Spiga M-G, Bishopric NH, Webster KA. A unique pathway of cardiac myocyte death caused by hypoxia–acidosis. J Exp Biol. 2004;207(18):3189–200. https://doi.org/10.1242/jeb.01109.
Harper AE, Miller RH, Block KP. Branched-chain amino acid metabolism. Annu Rev Nutr. 1984;4(1):409–54. https://doi.org/10.1146/annurev.nu.04.070184.002205.
Huang L, Li T, Liu Y-W, Zhang L, Dong Z-H, Liu S-Y, Gao Y-T. Plasma metabolic profile determination in young ST-segment elevation myocardial infarction patients with ischemia and reperfusion: ultra-performance liquid chromatography and mass spectrometry for pathway analysis. Chin Med J. 2016;129(9):1078.
Khan HA, Alhomida AS, Habib SS, Ola M, Khan A, Siddiqui NJ, Sobki SH, Al Madani H. Blood carnitine as a biomarker for acute myocardial infarction. Biomed Res. 2014;25:63–6.
Laborde CM, Mourino-Alvarez L, Posada-Ayala M, Alvarez-Llamas G, Serranillos-Reus MG, Moreu J, Vivanco F, Padial LR, Barderas MG. Plasma metabolomics reveals a potential panel of biomarkers for early diagnosis in acute coronary syndrome. Metabolomics. 2014;10(3):414–24.
Lu J, Chen B, Chen T, Guo S, Xue X, Chen Q, Zhao M, Xia L, Zhu Z, Zheng L. Comprehensive metabolomics identified lipid peroxidation as a prominent feature in human plasma of patients with coronary heart diseases. Redox Biol. 2017;12:899–907.
Maragos WF, Young KL, Altman CS, Pocernich CB, Drake J, Butterfield DA, Seif I, Holschneider DP, Chen K, Shih JC. Striatal damage and oxidative stress induced by the mitochondrial toxin malonate are reduced in clorgyline-treated rats and MAO-A deficient mice. Neurochem Res. 2004;29(4):741–6.
Misra MK, Sarwat M, Bhakuni P, Tuteja R, Tuteja N. Oxidative stress and ischemic myocardial syndromes. Med Sci Monit. 2009;15(10):RA209–19.
National CGCU (2013) Myocardial infarction with ST-segment elevation: the acute management of myocardial infarction with ST-segment elevation.
Naz S, Calderón ÁA, García A, Gallafrio J, Mestre RT, González EG, de Cabo CM, Delgado MCM, Balanza JÁL, Simionato AVC. Unveiling differences between patients with acute coronary syndrome with and without ST elevation through fingerprinting with CE-MS and HILIC-MS targeted analysis. Electrophoresis. 2015;36(18):2303–13.
Pouralijan Amiri M, Khoshkam M, Salek RM, Madadi R, Faghanzadeh Ganji G, Ramazani A. Metabolomics in early detection and prognosis of acute coronary syndrome. Clin Chim Acta. 2019;495:43–53. https://doi.org/10.1016/j.cca.2019.03.1632.
Rustom R, Wang B, McArdle F, Shalamanova L, Alexander J, McArdle A, Thomas CE, Bone JM, Shenkin A, Jackson MJ. Oxidative stress in a novel model of chronic acidosis in LLC-PK1 cells. Nephron Exp Nephrol. 2003;95(1):e13–23. https://doi.org/10.1159/000073019.
Senn T, Hazen SL, Tang WW. Translating metabolomics to cardiovascular biomarkers. Prog Cardiovasc Dis. 2012;55(1):70–6.
Surendran A, Aliani M, Ravandi A. Metabolomic analysis of human plasma during myocardial reperfusion injury: a non-targeted approach. J Am Coll Cardiol. 2018;71(11 Supplement):A12.
Ussher JR, Elmariah S, Gerszten RE, Dyck JR. The emerging role of metabolomics in the diagnosis and prognosis of cardiovascular disease. J Am Coll Cardiol. 2016;68(25):2850–70.
Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448.
Worley B, Powers R. Multivariate analysis in metabolomics. Current Metabolomics. 2013;1(1):92–107. https://doi.org/10.2174/2213235X11301010092.
Wu C, Chen HC, Chen ST, Chiang SY, Wu KY. Elevation in and persistence of multiple urinary biomarkers indicative of oxidative DNA stress and inflammation: toxicological implications of maleic acid consumption using a rat model. Plos One. 2017;12(10). https://doi.org/10.1371/journal.pone.0183675.
Xu X, Gao B, Guan Q, Zhang D, Ye X, Zhou L, Tong G, Li H, Zhang L, Tian J. Metabolomic profile for the early detection of coronary artery disease by using UPLC-QTOF/MS. J Pharm Biomed Anal. 2016;129:34–42.
Yao H, Shi P, Zhang L, Fan X, Shao Q, Cheng Y. Untargeted metabolic profiling reveals potential biomarkers in myocardial infarction and its application. Mol BioSyst. 2010;6(6):1061–70.
Yin X, de Carvalho LP, Chan MY, Li SFY. Integrated metabolomics and metallomics analyses in acute coronary syndrome patients. Metallomics. 2017;9(6):734–43.
Zhu M, Han Y, Zhang Y, Zhang S, Wei C, Cong Z, Du W. Metabolomics study of the biochemical changes in the plasma of myocardial infarction patients. Front Physiol. 2018;9:1017.
Acknowledgments
The authors thank all the participants who helped us in the collection of samples.
Funding
Not applicable
Author information
Authors and Affiliations
Contributions
YK and FG contributed to the collection of blood samples. OS contributed to the study design, experiment, statistical analysis, and manuscript writing. FDM contributed to the study design, experiment, and manuscript writing. GG contributed to the study design, metabolite profiling assay, statistical analysis, and manuscript writing. All authors contributed to the approval of the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gundogdu, G., Demirkaya Miloglu, F., Senol, O. et al. Investigation of the metabolic difference between ST-elevated myocardial infarction and non-ST-elevated myocardial infarction via LC/Q-TOF/MS/MS. J Anal Sci Technol 10, 31 (2019). https://doi.org/10.1186/s40543-019-0191-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40543-019-0191-3