Abstract
Background
Streptococcus pneumoniae is a significant global pathogen that colonises the nasopharynx of healthy children. Pneumococcal conjugate vaccines, which reduce nasopharyngeal colonisation of vaccine-type S. pneumoniae, may have broader effects on the nasopharyngeal microbiota; however, data are limited. In Fiji, nasopharyngeal carriage prevalence of S. pneumoniae and other colonising species differ between the two main ethnic groups. Here, we examined the association between the 7-valent pneumococcal conjugate vaccine (PCV7) and the nasopharyngeal microbiota of children in Fiji, including for each of the two main ethnic groups—indigenous Fijians (iTaukei) and Fijians of Indian descent (FID).
Method
The nasopharyngeal microbiota of 132 Fijian children was examined using nasopharyngeal swabs collected from 12-month-old iTaukei and FID children who were vaccinated (3 doses PCV7) or unvaccinated in infancy as part of a phase II randomised controlled trial. Microbiota composition was determined by sequencing the V4 region of the 16S rRNA gene. Species-specific carriage of S. pneumoniae, Haemophilus influenzae, Moraxella catarrhalis and Staphylococcus aureus was determined using real-time quantitative PCR. Associations between microbiota composition and other host and environmental factors were considered in the analysis.
Results
PCV7 had no overall impact on microbial diversity or composition. However, ethnic differences were observed in both diversity and composition with iTaukei children having higher relative abundance of Moraxella (p = 0.004) and Haemophilus (p = 0.004) and lower relative abundance of Staphylococcus (p = 0.026), Dolosigranulum (p = 0.004) and Corynebacterium (p = 0.003) compared with FID children. Further, when we stratified by ethnicity, associations with PCV7 could be detected: vaccinated iTaukei children had a lower relative abundance of Streptococcus and Haemophilus compared with unvaccinated iTaukei children (p = 0.022 and p = 0.043, respectively); and vaccinated FID children had a higher relative abundance of Dolosigranulum compared with unvaccinated FID children (p = 0.037). Children with symptoms of an upper respiratory tract infection (URTI) had a significantly different microbiota composition to children without symptoms. The microbiota composition of iTaukei children without URTI symptoms was most similar to the microbiota composition of FID children with URTI symptoms.
Conclusions
Associations between PCV7 and nasopharyngeal microbiota differed within each ethnic group. This study highlights the influence that ethnicity and URTIs have on nasopharyngeal microbiota.
Similar content being viewed by others
Background
Streptococcus pneumoniae (the pneumococcus) is a Gram-positive bacterium that causes a range of diseases, including otitis media, pneumonia and meningitis, and is a major cause of morbidity and mortality in children under 5 years of age worldwide [1]. Colonisation of the nasopharynx by S. pneumoniae is generally asymptomatic and is considered a precursor for pneumococcal disease [2, 3]. Pneumococcal conjugate vaccines (PCVs) target the pneumococcal capsular polysaccharide of common disease-causing serotypes, with current paediatric vaccine formulations targeting 10 or 13 out of over 90 known serotypes [4]. PCVs provide direct protection against infection and carriage. The reduction in vaccine-type carriage in vaccinees reduces transmission to unvaccinated individuals, thereby resulting in indirect (herd) effects [5,6,7]. In settings where PCVs have been introduced, serotype replacement has occurred, whereby serotypes not included in PCVs have become more prominent in carriage and disease [8, 9]. There is conflicting evidence about whether species replacement can also occur following pneumococcal vaccination, with mixed evidence for the common respiratory pathogens Haemophilus influenzae, Moraxella catarrhalis and Staphylococcus aureus [10,11,12,13]. Interactions with S. pneumoniae are thought to underpin vaccine-related changes in prevalence for these pathogens, with a negative interaction between S. pneumoniae and S. aureus, and generally positive associations between S. pneumoniae and H. influenzae, and S. pneumoniae and M. catarrhalis [14,15,16,17].
There is some evidence that PCVs may alter nasopharyngeal bacterial composition and diversity; however, findings have not been consistent across the four published studies [18,19,20,21]. Biesbroek et al. found that the 7-valent pneumococcal conjugate vaccine (PCV7) was associated with shifts in microbial composition and increases in bacterial diversity in Dutch children at 12, but not at 24, months of age [18]. In contrast, Feazel et al. found the 10-valent pneumococcal conjugate vaccine (PCV10) had no effect on the microbiome of Kenyan children (aged 12–59 months) 180 days after vaccination [19]. Two studies have been done in Swiss children. In children < 2 years of age with acute otitis media, PCV7 reduced the prevalence of commensal families [20]. The other study in healthy infants over the first year of life found that children vaccinated in the 13-valent pneumococcal conjugate vaccine (PCV13) era had higher microbial diversity and microbiota stability than children vaccinated in the PCV7 era; this was likely associated with the lower pneumococcal carriage prevalence in the PCV13 era [21].
Carriage prevalence of bacterial species can vary by geographic region and socio-economic status [22, 23]. Children in low- and middle-income countries, and susceptible populations in high-income settings, often have a high prevalence of pneumococcal colonisation, suggesting that the impact of vaccination on the microbiome could be different in these settings compared with children in developed countries [22].
There are two main ethnic groups in Fiji, indigenous Fijians (iTaukei) and Fijians of Indian descent (FID). The iTaukei have higher carriage prevalence of S. pneumoniae and higher burden of pneumococcal disease compared with FID [24, 25]. Previously, we conducted the Fiji Pneumococcal Project (FiPP), a randomised phase II vaccine trial that evaluated various schedules of PCV7 followed by the 23-valent pneumococcal polysaccharide vaccine as a booster at 12 months of age [26,27,28,29]. Results from FiPP at 17 months of age, and a long-term follow-up to the study, highlighted differences in carriage prevalence of S. pneumoniae, S. aureus, H. influenzae and M. catarrhalis between the two main ethnic groups [25, 30], suggesting they have distinct microbial profiles. We also found that the 23-valent pneumococcal polysaccharide vaccine given at 12 months of age had differential effects on long-term carriage of S. pneumoniae and S. aureus for the two ethnic groups [30].
Therefore, we hypothesise that PCVs may have differential effects on microbiota depending on the ethnicity of the child. To examine this directly, we used samples collected as part of the FiPP to determine the association between PCV and nasopharyngeal microbiota in Fijian children aged 12 months, including specific associations within each ethnic group.
Results
Overall, 144 samples (36 samples each from unvaccinated iTaukei children, PCV7 vaccinated iTaukei children, unvaccinated FID children and PCV7 vaccinated FID children), 4 sample repeats, 7 extraction controls and 5 PCR (no template) controls were sequenced resulting in a total of 62,038,435 sequence reads and an average of 387,740 reads per sample. Following sequence processing, 59% (36,344,642 total; 227,154 per sample) of these sequence reads remained for analysis. Twelve samples were excluded because they shared greater than 20% sequence read similarity with controls (n = 11) or had fewer than 50,000 sequence reads (n = 1). In the remaining 132 samples, we identified 4036 operational taxonomic units (OTUs) in total and 847 OTUs with unique taxonomic names. The most common genera (Dolosigranulum, Pseudomonas, Corynebacterium, Moraxella, Haemophilus, Streptococcus and Staphylococcus) accounted for at least 90% of all sequence reads in 118 out of the 132 (89%) of samples.
Study participant characteristics
Participant characteristics by ethnicity and vaccination status are shown in Table 1. There were few differences between unvaccinated and vaccinated children within each ethnic group, although among FID children, there were fewer males in the unvaccinated group compared with the vaccinated group and vaccinated children were slightly heavier. There was some evidence that fewer vaccinated FID children used antibiotics in the previous 2 weeks compared with unvaccinated FID children (0% vs. 13%). There was also some evidence that a greater number of unvaccinated iTaukei children were breastfed at the time of swab collection compared with vaccinated iTaukei children (79% vs. 56%). Pooled participant characteristics by both vaccination status and by ethnicity were also considered (see Additional file 1: Tables S1 and S2), with a higher proportion of unvaccinated children breastfed compared with vaccinated children (74% vs. 54%, p = 0.019). Mean weight was higher in iTaukei children compared with FID children (5023 g vs. 4382 g, p < 0.001) and more iTaukei children were exposed to cigarette smoke than FID children (55% vs. 18%, p = 0.038).
Overall association between PCV7 and nasopharyngeal microbiota
PCV7 vaccination had no significant association with the richness of nasopharyngeal microbiota (median number of OTUs in unvaccinated children 84 vs. 62 in vaccinated children, p = 0.137) or Shannon diversity (median of 1.36 in unvaccinated children vs. 1.28 in vaccinated children, p = 0.115) (see Additional file 2: Figure S1a and S1b). We explored differences in overall microbial composition between unvaccinated and vaccinated children, using Jaccard dissimilarity (both abundance-based and binary-based) and using non-metric multidimensional scaling (nMDS) plots as an ordination method. Overall, vaccination appeared to have little impact on the microbial composition of the nasopharynx regardless of whether abundance-based (Fig. 1a, p = 0.668) or binary-based (Fig. 1b, p = 0.893) dissimilarity was considered. Looking specifically at the seven most common genera (Table 2), PCV7 was only associated with changes to the relative abundance of Streptococcus (p = 0.030), with a lower relative abundance of Streptococcus in vaccinated children (median 0.59%) compared with unvaccinated children (median 1.02%). The random forest model considering the association between PCV7 vaccination and microbial composition was not significant (p = 0.836). As the 16S sequencing analysis was generally limited to genus level, we examined carriage prevalence and density of the common respiratory pathogens S. pneumoniae, H. influenzae, M. catarrhalis and S. aureus using species-specific real-time quantitative PCR (qPCR), finding no differences between unvaccinated children and vaccinated children (Additional file 3:Figure S2).
nMDS of the dissimilarity in microbial composition using both abundance-based and binary-based measures by vaccination status (a and b, respectively), by ethnicity (c and d, respectively), in iTaukei children by vaccination status (e and f, respectively) and in FID children by vaccination status (g and h, respectively). The centre points represent the mean of each group and the lines represent distance from the mean for each sample. p values calculated using PERMANOVA. Stress values for figures are 0.158 (a and c), 0.216 (b and d), 0.154 (e), 0.218 (f), 0.149 (g) and 0.215 (h)
Ethnic differences
We next examined difference in nasopharyngeal microbiota by ethnic group. While the ethnicity of a child did not influence OTU richness (median number of OTUs in iTaukei children of 68 vs. 72 in FID children, p = 0.436), it did have an influence on Shannon diversity. OTUs were more evenly spread in iTaukei children compared with FID children (median Shannon diversity index of 1.33 vs. 1.07, p = 0.006) (see Additional file 2: Figure S1c and S1d). The overall microbial composition of the nasopharynx was significantly different between iTaukei and FID children using both abundance-based (p < 0.001) and binary-based measures (p < 0.001) (Fig.1c and d). Of the most common genera (Table 2), iTaukei children had higher relative abundance of Moraxella and Haemophilus and lower relative abundance of Staphylococcus, Dolosigranulum and Corynebacterium. The random forest model also showed an association between ethnicity and microbial composition (p < 0.001). The most important OTUs in the random forest model included Corynebacterium, Staphylococcus and Moraxella, as well as less abundant OTUs Helcococcus, Acinetobacter and Prevotella (Fig. 2a). This Helcococcus OTU was more prevalent in iTaukei children compared with FID children (78% in iTaukei vs. 52% in FID children, p = 0.003) and was also present at a higher relative abundance (median relative abundance of 0.4% in iTaukei children vs. 0.001% in FID children, p < 0.001). By species-specific qPCR, the carriage prevalence of S. pneumoniae, H. influenzae and M. catarrhalis was higher in iTaukei children than FID children (69% vs. 26%, 69% vs. 28% and 91% vs. 37%, respectively, all p < 0.001). The carriage prevalence of S. aureus was similarly low in iTaukei children (4%) and FID children (6%). No differences in species-specific density between ethnic groups were observed (Additional file 3: Figure S3).
Association between PCV7 and nasopharyngeal microbiota within each ethnic group
Given the differences in microbiota composition between ethnic groups, we examined the association between PCV7 and nasopharyngeal microbiota after stratifying by ethnicity. There were no significant differences in OTU richness or Shannon diversity between unvaccinated and vaccinated children within either ethnic group (iTaukei children: median of 84 OTUs and 1.36 Shannon diversity in unvaccinated children vs. 62 OTUs and 1.28 Shannon diversity in vaccinated children, p = 0.098 and 0.115, respectively; FID children: median of 70 OTUs and 1.11 Shannon diversity in unvaccinated children vs. 71 OTUs and 1.00 Shannon diversity in vaccinated children, p = 0.095 and 0.662, respectively) (see Additional file 2: Figure S1e and S1f). nMDS analyses suggested PCV7 had little association with the nasopharyngeal microbiota of iTaukei children using both abundance-based (p = 0.375) and binary-based (p = 0.155) measures (Fig. 1e and f). There was some evidence that PCV7 was associated with nasopharyngeal microbial composition of FID children using an abundance-based measure (p = 0.05) but not a binary-based measure (p = 0.479) (Fig. 1g and h), suggesting that PCV7 may impact the relative abundance, but not the presence, of OTUs in FID children. Random forest models showed no association between microbial composition and vaccination within either ethnic group (p = 0.952 in Taukei children and p = 0.512 in FID children).
In iTaukei children, unvaccinated children had higher relative abundance of Streptococcus compared with vaccinated children (Table 2). Similarly, unvaccinated iTaukei children had a higher relative abundance of Haemophilus compared with vaccinated iTaukei children. No other differences in the most common genera between unvaccinated and vaccinated iTaukei children were seen.
In FID children, those that were unvaccinated had lower relative abundance of Dolosigranulum compared with children who received PCV7 (Table 2). No other PCV7-associated differences in common genera were observed in FID children.
Using species-specific qPCR assays, we examined carriage prevalence and density within each ethnic group for S. pneumoniae, H. influenzae, M. catarrhalis and S. aureus comparing unvaccinated and vaccinated children (Additional file 3: Figure S4). Within each ethnic group, there were no significant differences in carriage prevalence or density by vaccination status for any of the four bacterial species examined.
Symptoms of an upper respiratory tract infection
We examined the association between other factors (seasonality, exposure to cigarette smoke, antibiotic use in the previous 2 weeks, year of swab collection, sex, current breastfeeding status and symptoms of an upper respiratory tract infection (URTI)—either cough or runny nose) and overall microbiota composition (Additional file 1: Table S1). Only the presence of symptoms of an URTI had a significant impact on microbial composition. In the random forest model for URTI symptoms (p = 0.012), the most important OTUs were Staphylococcus, an unclassified Pasteurellaceae OTU, Haemophilus, Moraxella, Veillonella and Helcococcus (Fig. 2b). Compared with children without URTI symptoms, children with URTI symptoms had higher relative abundance of Moraxella, Haemophilus and Streptococcus and lower relative abundance of Dolosigranulum and Corynebacterium (Table 3).
Interestingly, not only did children with symptoms of an URTI have significantly different microbiota compared with children without symptoms, but the microbiota composition of iTaukei children without URTI symptoms was most similar to the microbiota composition of FID children with URTI symptoms (Fig. 3). When analysis of the most common genera was stratified by ethnicity, the relative abundance in Haemophilus was higher in iTaukei children with URTI symptoms compared to those without symptoms (Additional file 1: Table S2). No other significant differences were observed, but trends were consistent in both ethnic groups and with the overall results shown in Table 3.
nMDS of the dissimilarity in microbial composition by ethnicity and symptoms of an upper respiratory tract infection using abundance-based (a) and binary-based (b) measures. Shown are iTaukei children, with (light blue) or without (dark blue) symptoms of an upper respiratory tract infection, and FID children, with (light brown) or without (dark brown) symptoms of an upper respiratory tract infection. The centre points represent the mean of each group and the lines represent distance from the mean for each sample. There was a significant difference between the four groups (p = 0.001), p value calculated using PERMANOVA. Stress values for figures are 0.158 (a) and 0.216 (b)
Microbial composition and dominant taxa
We next looked at microbial composition in the samples by generating a heatmap and clustering samples based on their composition using a Jaccard distance matrix (Fig. 4). Most samples were dominated by one or two taxa with the following clusters identified by their dominant taxa (Fig. 3): Cluster 2 (n = 36) dominated by Moraxella; Cluster 3 (n = 22) dominated by Dolosigranulum; Cluster 4 (n = 34) dominated by Dolosigranulum; Cluster 5 (n = 16) dominated by Haemophilus and Corynebacterium; and Cluster 6 (n = 20) dominated by Pseudomonas. Cluster 1 contained four samples that did not fit into any of the other clusters.
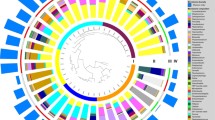
Relative abundance (%) heatmap including all 132 samples. Sample clustering is shown on the left-hand side. Taxa with a relative abundance above 30% in at least one sample are shown at the bottom. Where multiple OTUs are from the same genus ‘.1’ has been used for subsequent OTUs. ‘Others’ represents the remaining taxa in each sample. Clusters for samples have been coloured to show clustered groups: Cluster 1 (shown in dark blue), Cluster 2 (shown in green) dominated by Moraxella, Cluster 3 (shown in yellow) dominated by Dolosigranulum, Cluster 4 (shown in purple) dominated by Dolosigranulum and Corynebacterium, Cluster 5 (shown in orange) dominated by Haemophilus, and Cluster 6 (shown in grey) dominated by Pseudomonas. The vaccination status (unvaccinated—blue; vaccinated—red), ethnicity (iTaukei—turquoise; FID—gold) and whether the child had any symptoms of an upper respiratory tract infection (no URTI—light blue; URTI—brown) are shown in bars on the right of the heatmap
Associations between clusters and host factors were considered, with several clusters associated with ethnicity and symptoms of an URTI. Compared with iTaukei children, FID children were more likely to have a cluster 4 microbiota profile (40% FID children vs. 12% iTaukei children, p < 0.001) and less likely to have a cluster 2 microbiota profile (17% FID children vs. 37% iTaukei children, p = 0.011). Cluster 4 was associated with a higher proportion of children without URTI symptoms (31% no URTI children vs. 13% URTI children, p = 0.030). There was also some evidence that cluster 5 and cluster 2 were associated with symptoms of an URTI (9% no URTI children vs. 21% URTI children, p = 0.078; and 23% no URTI children vs. 38% URTI children, p = 0.086, respectively).
Owing to concern that Pseudomonas was a potential contaminant, we cultured eight samples where Pseudomonas comprised of > 90% of sequence reads. We detected Pseudomonas on selective agar for six of the eight samples (two samples had no growth), which were identified as P. fluorescens (n = 3) and P. fragi (n = 3) by MALDI-TOF MS.
Bacterial interactions
We examined the relationships between the 100 most common OTUs using the network inference tool, SparCC (Fig. 5). Of particular note for the correlation network, the predominant Streptococcus OTU had positive relationships with two of the Moraxella OTUs (r = 0.32, p < 0.001 and r = 0.34, p < 0.001) and the predominant Haemophilus OTU (r = 0.36, p < 0.001); and a negative relationship with Pseudomonas (r = 0.31, p < 0.001). There was no significant interaction between Streptococcus and Dolosigranulum (r = 0.03, p = 0.379).
Network correlation map based on the 100 most common OTUs considering samples from all 132 children. Shown are SparCC correlations with p value < 0.05 and r > 0.3. The main OTUs from the top seven genera are shown by larger node size. Colours of nodes are grey except for the seven most common genera. The edge (connecting line) colour represents a positive (green solid lines) or negative (red dashed lines) correlation between two OTUs, with thicker lines representing stronger correlations. Only those correlations with genus-level classification are shown and are labelled with the OTU genus
Discussion
There are limited data on the impact of pneumococcal vaccination on respiratory microbiota, particularly from children in low- and middle-income countries. We investigated the association between pneumococcal vaccination and nasopharyngeal microbiota in 12-month-old Fijian children approximately 8.5 months after their third dose of PCV7. Given that the two ethnic groups in Fiji have marked differences in pneumococcal carriage and disease prevalence, we examined the associations with pneumococcal vaccination by ethnicity.
We found little evidence of a broader association between PCV7 and nasopharyngeal microbiota, beyond reducing the relative abundance of Streptococcus in vaccinated children compared with unvaccinated children. Our findings were consistent with a study in Kenyan children aged 12–59 months, where no differences were observed 4 months after vaccination [19]. Some effects of pneumococcal vaccination have been observed in studies in Swiss children with otitis media aged < 2 years, where vaccination reduced commensals such as Streptococcaeae (excluding pneumococci) and Corynebacteriaeae [20], and in Dutch children aged 12 months, where vaccinated children had increased absolute abundance of Haemophilus and Staphylococcus as well as Actinomyces, Rothia, Neisseria and Veillonella [18]. There are some notable differences between these studies and ours, particularly with regard to vaccine formulation and schedule and age at which microbiota was assessed. These differences include the third dose of PCV in our study given to children at 14 weeks of age compared to 11 months of age in the Netherlands study and the use of the 10-valent PCV in the Kenyan study rather than 7-valent PCV used in our study. Additionally, in some cases, the laboratory methods (such as the sequencing platform, the 16S rRNA region sequenced and the DNA extraction method used) differed between studies which may contribute to some differences in findings. Sequencing depth also varies between studies, and some studies use a subsampling approach for analysis. We conducted an exploratory analysis using subsampling and found no observable differences (data not shown) and therefore did not use subsampling in our analyses. We cannot exclude the possibility that other methodological approaches, such as the use of other DNA extraction methods, would yield slightly different results. However, available data suggests that any PCV effects on nasopharyngeal microbiota may occur soon after vaccination and may be transient.
Our previous studies in Fiji [25, 30] found significant differences in bacterial carriage between iTaukei and FID children but only examined a limited number of bacterial species. Here, we found substantial differences in the entire nasopharyngeal microbiota based on ethnicity. Ethnicity influenced both microbial community structure, with higher Shannon diversity in iTaukei children, and community composition, with differences in the relative abundance of five of the seven most common genera. Of the participant characteristics collected, only weight and exposure to household cigarette smoking differed between the two ethnic groups, and in this study neither of these had a significant association with microbial composition. There are several genetic, environmental and social factors which may contribute to the observed ethnic differences in the microbiota [31]. Differences in genes associated with immunity, such as beta-defensin and pattern recognition receptors have been associated with differences in the nasopharyngeal microbiota and susceptibility to bacterial infections [32, 33]. Polymorphisms, particularly in immunity-related genes, have been linked to microbial community composition, and patterns of polymorphisms have shown to differ by geographic and ethnic origin [34, 35]. There is some evidence that Helcococcus, highlighted as one of the most significant OTUs in the random forest model for ethnicity, is associated with the presence of chemokine CXL8 [36], supporting a role for immune factors in the observed ethnic differences.
The 2004 Fiji National Nutrition survey revealed several dietary and lifestyle factors that differed between the two ethnic groups including differences in consumption of white meat (e.g. chicken) and preserved foods, as well as levels of physical activity and cigarette use [37]. There is some evidence that diet and composition of intestinal microbiota may be determinants of respiratory microbiota [38, 39]; however, information on diet (apart from breastfeeding status) was not collected in our study.
Other household characteristics like social contact and/or environmental factors may also be important in Fiji. Recently, we undertook a social contact survey in Fiji, finding that iTaukei children have a greater frequency of physical contacts than FID children [40]. There is evidence that social contact can influence microbiota [41] but the relative importance of social contact compared with other factors as a determinant of microbiota is unknown. In Fiji, the two main ethnic groups have similar household income [30] so it is unlikely that socio-economic status, a known risk factor for pneumococcal carriage [23], contributes to the observed differences in nasopharyngeal bacteria between iTaukei and FID children. Given the complexity and interconnected nature of factors that can determine microbiota, it is likely multiple factors are involved in the underlying differences in microbial composition between ethnic groups in Fiji.
We next examined the associations with PCV7 after stratifying by ethnicity. In iTaukei children, PCV7 vaccination was associated with reduced relative abundance of both Streptococcus and Haemophilus, without associations with other common genera. We noted a positive relationship between Streptococcus and Haemophilus, which may underpin an indirect reduction in Haemophilus (i.e. through a direct vaccine-induced reduction in Streptococcus). However, there was no evidence that vaccination reduced the carriage prevalence or density of S. pneumoniae or H. influenzae. While PCVs reduce carriage prevalence of vaccine serotype S. pneumoniae, they generally have no effect on overall S. pneumoniae carriage prevalence due to serotype replacement by non-vaccine serotypes [42, 43], and so the lack of an overall association in this study with S. pneumoniae carriage prevalence is not unexpected. We also saw no association between PCV7 and S. pneumoniae carriage density despite the observed difference in relative abundance of Streptococcus. However, as the density of S. pneumoniae was an absolute measure rather than relative, it is possible that differences in absolute abundance between unvaccinated and vaccinated children may explain the discrepancy between the two sets of results. Consistent with this, Bisbroek et al. [18] found that the absolute abundance of total microbiota was much higher in vaccinated children compared with controls at 24 months of age.
In contrast to the effect in iTaukei children, PCV7 was not associated with changes to the relative abundance of Streptococcus, nor S. pneumoniae carriage prevalence or density, in FID children. However, the relative abundance of Dolosigranulum was significantly higher in vaccinated FID children compared with unvaccinated FID children. Dolosigranulum has been reported to have a negative relationship with S. pneumoniae [44], and as such, although we noted no association between PCV7 and Streptococcus and no direct interaction between Streptococcus and Dolosigranulum, our findings may have been a result of earlier interactions not captured in our sampling. For example, it is plausible that pneumococcal vaccination may have had a transient effect on Streptococcus directly after vaccine administration, which may also have indirectly increased the relative abundance of Dolosigranulum, in vaccinated FID children. Microbiota profiles containing Dolosigranulum are more stable than those containing Streptococcus [45], and therefore it is possible that an effect may have been maintained for Dolosigranulum but not Streptococcus.
The presence of URTI symptoms had a significant association with nasopharyngeal microbiota. In children with symptoms of an URTI, the relative abundances of Streptococcus, Moraxella and Haemophilus was higher and the relative abundance of Dolosigranulum and Corynebacterium was lower, compared with children without URTI symptoms. Interestingly, when we looked at community composition by ethnicity and URTI symptoms, we found that the microbiota of FID children with URTI symptoms was similar to the microbiota of iTaukei children without URTI symptoms, although the presence of URTI symptoms shifted microbiota profiles in a similar direction for both ethnic groups. The association with URTI symptoms and Moraxella and Haemophilus is perhaps unsurprising given that there are known respiratory pathogens within the genera which have been linked to URTIs [46, 47]. Dolosigranulum and Corynebacterium have been linked with a healthy nasopharyngeal microbiome, and the presence of Dolosigranulum and Corynebacterium have been associated with a decreased risk of otitis media [44] as well as decreased episodes of respiratory tract infections [45, 48].
A limitation of this study is that viruses were not examined, and URTI symptoms may be related to viral infection. De Steenhuijsen Piters et al. [49] found that respiratory syncytial virus infection and hospitalisation was associated with Streptococcus and Haemophilus in the nasopharyngeal microbiota in children < 2 years of age. In adults, rhinovirus decreased α-diversity and altered the relative abundance of the genera Neisseria and Propionibacterium [50].
Contamination can be a significant problem in microbiome studies, particularly for low biomass samples [51]. We sequenced all the PCR products from the extraction and PCR controls, as despite taking several measures to reduce contamination and repeating PCRs with no template added, a PCR product was present in all our controls. Contamination is difficult to eliminate in microbiome studies due to the presence of bacterial components in purified water, DNA extraction kits and PCR reagents [51, 52]. The OTU composition of the controls was substantially different to the samples included in our analysis, giving confidence in our findings. Furthermore, most of the abundant genera in our study have been reported as predominant genera in other studies [20, 53]. The exception is Pseudomonas, which is not commonly found in the nasopharynx, but has been identified as a potential contaminant in several studies [51, 54]. In our study, when Pseudomonas was found, it was generally present at a high relative abundance in participant samples and at a low relative abundance in the ‘typical’ control profile (< 1% in 10 out 12 controls). Samples containing Pseudomonas were distributed across all seven rounds of DNA extractions. We consider it unlikely that Pseudomonas is a contaminant in our study, as viable Pseudomonas from two different species were cultured from six out of eight samples for which Pseudomonas accounted for > 90% of the relative abundance. Nevertheless, when we removed these eight samples from our analysis it did not affect the conclusions of our study. Additionally, given that for other Pseudomonas species such as P. aeruginosa acquisition increases in warmer seasons and tropical climates [55, 56], it is plausible that nasopharyngeal carriage of Pseudomonas may occur in Fiji.
A key strength of this study is that samples were obtained from a randomised control trial in a setting where PCV has not been introduced. Additionally, PCV7 was not available on the private market when the samples were collected, therefore herd effects are unlikely to impact unvaccinated participants in our study. This means that participants would not be subject to any indirect effects of PCV, and samples were collected during the same time period and from children in a similar setting. Limitations are that the samples were tested following approximately 10 years of storage at − 80 °C, that PCV7 has been replaced by higher valency vaccines (PCV10 and PCV13 are currently in use globally) and that only a single time point was tested. Long-term frozen storage may impact the quality and quantity of DNA recovered from swab samples and has the potential to have an effect on the microbial community structure [57]. However, any effects related to storage are likely to be consistent between groups and therefore would not affect overall conclusions. One limitation of our study is the relatively small sample size, which was partly due to ensuring that there were even numbers of children from each ethnic group represented. Microbiota were not examined at baseline, and groups were not randomised by baseline microbiota. Given the small sample size, it is possible that baseline microbiota may have varied between groups. As such, this study was focussed on exploring associations between PCV7 and nasopharyngeal microbiota rather than inferring direct causality. Given the effect that a small sample may have on some analyses, and potential issues of multiplicity, results should be interpreted with caution. Although PCV7 affects fewer serotypes as PCVs with higher valency, the mechanism of protection is the same; therefore, any trends observed are likely to be similar to those observed for PCV10 or PCV13.
Conclusion
Overall, we found no significant wider association between PCV and nasopharyngeal microbiota. These results suggest that the majority of replacement that occurs following the elimination of vaccine type pneumococci from the nasopharynx is due to non-vaccine type pneumococci, rather than other bacterial species. However, we found distinct microbial profiles by ethnic group, as well as evidence that the associations with pneumococcal vaccination varied between ethnic groups. Pneumococcal vaccination reduced the relative abundance of Streptococcus and Haemophilus in iTaukei children and increased the relative abundance of Dolosigranulum in FID children. Given the association of Streptococcus and Haemophilus with URTIs and the association of Dolosigranulum with a healthy microbiome, we found that pneumococcal vaccination had positive associations with microbiota in both ethnic groups.
Methods
Study design
The swabs used in this study were collected as part of the FiPP, a phase II vaccine trial in Suva, Fiji [26,27,28,29]. Details of selection criteria, randomisation procedures and results of the vaccine trial are reported elsewhere [26,27,28,29] with key details of trial design found in Russell et al. [27]. In brief, healthy infants were recruited from three participating health centres and were stratified by ethnicity and randomised into groups using a computer-generated list. Participant information was collected by nurses at enrolment (and at swab collection) from the parent/guardian; this included demographic information (e.g. date of birth, sex and ethnicity) and information on known risk factors for pneumococcal colonisation (e.g. exposure to household cigarette smoke, antibiotic use in the previous two weeks and symptoms of an URTI). In this study, we used a total of 144 nasopharyngeal swabs, 36 randomly selected from each of the four groups (iTaukei and FID children; who were either unvaccinated or vaccinated with three doses of PCV7 at 6, 10 and 14 weeks of age). Swabs were collected between 2005 and 2007 from FiPP participants at 12 months of age (prior to 23-valent pneumococcal polysaccharide vaccine administration) and stored and transported as previously described [26]. In brief, buffered cotton swabs (aluminium shaft-buffered; Sarstedt, Australia) were used to sample the nasopharynx for 5 s, before being placed in a sterile cryovial (Simport, Canada) containing 1 ml of skim milk tryptone glucose glycerol (STGG) medium. After chilled transport to the Colonial War Memorial Hospital in Suva, Fiji, the swabs were stored at − 70 °C. Swabs were transported on dry ice to the Pneumococcal Research Laboratory at MCRI in Melbourne, Australia, and stored at − 80 °C.
DNA extractions
DNA extractions were performed in batches of 23 samples (total of seven batches) plus a negative control (empty sterile 1.5-ml microfuge tube) using the QIamp DNA Mini Kit (Qiagen). In a biohazard class II cabinet, 200 μl of swab STGG was aliquoted into a sterile 1.5-ml microfuge tube for each sample. These aliquots were then spun in a centrifuge for 5 min at 6000×g to create a pellet. Pellets were placed at − 80 °C for 15 min to promote cell lysis. Following the freeze/thaw step, 200 μl of enzymatic lysis buffer (containing 36.25 mM phosphate buffer, 1 mg/ml lysozyme, 0.075 mg/ml mutanolysin and 2 mg/ml proteinase K) was added, followed by incubation at 56 °C for 45 min. An additional 20 μl of proteinase K (20 mg/ml, Qiagen) was added before further incubation at 56 °C for 10 min. 4 μl of RNase A (at 100 μg/ml, Qiagen) was added and mixed gently at room temperature for 2 min before adding 200 μl of buffer AL. Samples were then incubated at 70 °C for a further 10 min to complete cell lysis. Extractions were then performed according to the manufacturer’s instructions with DNA eluted in 100 μl of Buffer AE and DNA stored at − 30 °C.
16S rRNA gene PCR
The V4 region of the 16S rRNA gene was amplified by PCR using a primer pair which included the Illumina-specific adapter sequences (Table 4). Each PCR was performed using 1 μM of each primer (HPLC purified in liquid at 100 μM concentration, Sigma-Aldrich, Australia), 1X Phusion Green Hot Start II High-Fidelity PCR Master Mix (Thermo Fisher Scientific) and 10 μl of extracted gDNA in a 50-μl reaction. Each PCR run included a no template control which was sequenced along with extraction controls. Following an initial denaturation step at 98 °C for 30 s, there were 35 cycles of denaturation at 98 °C for 5 s, annealing at 65 °C for 20 s and elongation at 72 °C for 15 s, followed by a final elongation step at 72 °C for 5 min. PCR products were then stored at − 20 °C. PCR products from the V4 PCR were purified using AMPure XP beads and following the Illumina ‘16S Metagenomic Sequencing Library Preparation’ protocol (Part # 15044223 Rev. B). PCR product sizes and concentrations were measured by running samples on a TapeStation System (Agilent Technologies) using the D1000 ScreenTape System (Agilent Technologies). MiSeq sequencing was performed by the Translational Genomics Unit at MCRI, Melbourne, Australia. Initially, a sequencing run of 22 samples (plus the extraction control and the no template PCR control) was performed, with the remaining samples and controls then sequenced in two batches. As the sequencing depth varied between the initial sequencing run and the two batches performed later, four of the initial 22 samples were also included in the later sequencing batches to check results that did not differ by sequencing depth.
The V4 purified DNA was prepared for sequencing using the Illumina ‘16S Metagenomic Sequencing Library Preparation’ protocol (Document # 15044223 Rev. B). The libraries were sequenced using Illumina MiSeq V3 reagent kits (2 × 300 bp) on the MiSeq platform (Illumina).
Sequence processing
Raw sequences were processed and classified using MOTHUR (version 1.35.1) [58] following the MiSeq SOP (http://www.mothur.org/wiki/MiSeq_SOP; accessed 27 July, 2015) and Silva nr (v119) classification database [59]. Sequences with read lengths falling outside a 225–325 base pairs (bp) range, any ambiguous bases and > 8 bp homopolymers were removed. Sequences were aligned across the region spanning the forward and reverse primers, and any overhang was trimmed. Chimeric sequences were identified and removed using UCHIME [60] and sequences were clustered into OTUs at 97% similarity to the genus level. OTUs for which > 33% of sequence reads were from controls were removed from count data to reduce background signal. Samples which had less than 50,000 sequence reads were removed from further analysis, as were any samples that shared greater than 20% identity (based on relative abundance) to the controls (both extraction and PCR) in downstream analysis as these were considered to have amplified little, if any, bacterial DNA from the swab.
Species-specific qPCR
Species-specific qPCR assays were used to detect, and determine the carriage densities, of the respiratory pathogens S. pneumoniae, Staphylococcus aureus, Moraxella catarrhalis and Haemophilus influenzae [25]. Two duplex assays were performed using primers (Sigma-Aldrich), probes (Eurogentec) and concentrations as described previously [30]. Samples were run in duplicate wells using 2 μl of extracted DNA and Brilliant III Ultra-Fast qPCR Master Mix (Agilent Technologies). Standard curves for each species were prepared using a dilution series of DNA extracted from S. pneumoniae ATCC 6305, H. influenzae F412, M. catarrhalis ATCC 8176 and S. aureus ATCC 29213. Duplex qPCRs were performed on a Stratagene Mx3005 instrument with 40 cycles of 95 °C for 20 s and 60 °C for 20 s after an initial activation of 95 °C for 3 min. Results were analysed using MxPro™ software (Stratagene) and genomic equivalents per millilitre determined for each sample.
Pseudomonas culture
Eight samples (FM052, FM108, FM029, FM001, FM004, FM129, FM057, FM020) which contained Pseudomonas at a relative abundance > 90% were cultured on cephaloridine fucidin cetrimide (CFC) agar (Oxoid, Thermo Fisher) at 28 °C for 48 h. Isolated colonies were then confirmed as Gram-negative bacilli that were oxidase positive and identified by MALDI-TOF MS using the Vitek® MS system (bioMérieux).
Analyses
Analyses were conducted and graphs were generated using GraphPad Prism version 7.02 unless otherwise stated. P < 0.05 was considered statistically significant. Relative abundance was defined as the percentage of total sequence reads from an individual sample that were from the specific OTU, genus or family being considered. Relative abundance (%) data was used as input for analyses unless otherwise stated.
For comparing carriage prevalence from the species-specific qPCR data, Fisher’s exact test was used. For comparing carriage densities from the species-specific qPCR, data were first checked for normality using the D’Agostino & Pearson normality test. For normally distributed data, an unpaired t test was used. Where data were not normally distributed, the Mann-Whitney test was used.
Alpha-diversity (i.e. within sample diversity) was determined through OTU richness (number of OTUs) and Shannon diversity index. Alpha-diversity measures were calculated using R Studio (version 3.2.4) and the ‘diversity’ function of the vegan package (version 2.3-5) [61]. Richness calculations were rarefied to account for differences in sequencing depth, as greater sequencing depth increases the number of OTUs or richness. Rarefaction was done by subsampling 50,000 sequence reads (based on the lowest number of reads—53,616 reads) using the ‘rarefy’ function in vegan package. Beta-diversity (i.e. between sample diversity) was calculated using a Jaccard distance matrix (‘vegdist’ function, R vegan package) using both abundance-based (default setting) and binary-based (binary = TRUE) measures with nMDS plots (‘metaMDS’ function, vegan package) as the ordination method. Permutational multivariate analysis of variance (PERMANOVA, ‘adonis’ function, R vegan package) was used to calculate significant differences in microbial composition between groups.
A heatmap of microbial composition in all samples was generated in R using the following packages: vegan, RColorBrewer (version 1.1-2), gplots (version 3.0.1) and dendextend (version 1.7.0). Samples and OTUs were clustered using the ‘hclust’ function and the average linkage method based on a Jaccard distance matrix. For generating the heatmap, OTUs were filtered to only those with a relative abundance above 30% in at least one sample. Clusters were characterised using the ‘cascadeKM’ function (vegan package) with the default ‘calinski’ criterion (Calinski-Harabasz [62]) and coloured using the ‘color_branches’ function (dendextend package).
To examine co-presence or mutual-exclusion of the most abundant OTUs, a correlation network was generated using count data and the default SPARCC [63] parameters in MOTHUR after filtering to the top 100 most abundant OTUs. The SPARCC data was then formatted and analysed in R. Only correlations with a significance of p < 0.05, a correlation r > 0.3 and OTUs with a classification to the genus level were included in the network. Networks were visualised and edited using Cytoscape software version 3.3.0 [64].
Random forest models were used for each risk factor to rank OTUs on their ability to discriminate between groups (see Additional file 4: Table S4). The random forest analysis was done in RStudio using R packages ‘randomForest’ version 4.6-14, ‘pylr’ version 1.8.4 and ‘rfUtilities’ version 2.1-3. Using relative abundance data, rare OTUs that were zero in greater than 95% of samples were removed, data were then normalised back to 100%. Each risk factor was analysed in separate models using the ‘randomForest’ and ‘rf.significance’ functions. Each model used the parameters ‘ntree = 501’, ‘importance = TRUE’ and ‘proximities = TRUE’, and significance was calculated using 500 permutations. The mean decrease in accuracy or variable importance was examined in each model for the top 10 OTUs.
Multivariable linear regression (Additional file 5 and Additional file 6) was done in RStudio using the ‘lm’ function (base ‘stats’ package in R, version 3.2.4) for looking at specific associations with vaccination and ethnicity on pooled genus data or α-diversity measures. For each model, an interaction term between vaccination and ethnicity was considered and Akaike information criterion was used to determine whether inclusion of the interaction term was appropriate. As well as vaccination status and ethnicity, final models included the following risk factors: symptoms of an URTI (such as runny nose or cough), breastfeeding status, household exposure to cigarette smoke, the year the swab was collected, season of swab collection, antibiotic use in the previous two weeks and sex of the child. Each model also included the sequence count for each sample to account for potential differences in sequencing depth between samples. Checks of the normality of residuals were performed using the ‘plot’ function in R, in cases where residuals were not normal, data were log transformed.
Availability of data and materials
The datasets generated and analysed during the current study are available in the NCBI Sequence Read Archive, accession number PRJNA493513 (https://www.ncbi.nlm.nih.gov/sra/PRJNA493513).
Abbreviations
- FID:
-
Fijians of Indian descent
- FiPP:
-
Fiji Pneumococcal Project
- nMDS:
-
Non-metric multidimensional scaling
- OTU:
-
Operational taxonomic unit
- PCV:
-
Pneumococcal conjugate vaccine
- qPCR:
-
Real-time quantitative PCR
- URTI:
-
Upper respiratory tract infection
References
O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902.
Bogaert D, de Groot R, Hermans PWM. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4:144–54.
Simell B, Auranen K, Kayhty H, Goldblatt D, Dagan R, O’Brien KL, et al. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines. 2012;11:841–55.
Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, et al. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev. 2015;28:871–99.
Cohen R, Cohen JF, Chalumeau M, Levy C. Impact of pneumococcal conjugate vaccines for children in high- and non-high-income countries. Expert Rev Vaccines. 2017;16:625–40.
Tsaban G, Ben-Shimol S. Indirect (herd) protection, following pneumococcal conjugated vaccines introduction: a systematic review of the literature. Vaccine. 2017;35:2882–91.
Davis SM, Deloria-Knoll M, Kassa HT, O’Brien KL. Impact of pneumococcal conjugate vaccines on nasopharyngeal carriage and invasive disease among unvaccinated people: review of evidence on indirect effects. Vaccine. 2013;32:133–45.
Feikin DR, Kagucia EW, Loo JD, Link-Gelles R, Puhan MA, Cherian T, et al. Serotype-specific changes in invasive pneumococcal disease after pneumococcal conjugate vaccine introduction: a pooled analysis of multiple surveillance sites. PLoS Med. 2013;10:e1001517.
Gladstone RA, Jefferies JM, Tocheva AS, Beard KR, Garley D, Chong WW, et al. Five winters of pneumococcal serotype replacement in UK carriage following PCV introduction. Vaccine. 2015;33:2015–21.
Reiss-Mandel A, Regev-Yochay G. Staphylococcus aureus and Streptococcus pneumoniae interaction and response to pneumococcal vaccination: myth or reality? Hum Vaccin Immunother. 2016;12:351–7.
Dunne EM, Smith-Vaughan HC, Robins-Browne RM, Mulholland EK, Satzke C. Nasopharyngeal microbial interactions in the era of pneumococcal conjugate vaccination. Vaccine. 2013;31:2333–42.
Spijkerman J, Prevaes SM, van Gils EJ, Veenhoven RH, Bruin JP, Bogaert D, et al. Long-term effects of pneumococcal conjugate vaccine on nasopharyngeal carriage of S. pneumoniae, S. aureus, H. influenzae and M. catarrhalis. PLoS ONE. 2012;7:e39730.
van Gils EJ, Hak E, Veenhoven RH, Rodenburg GD, Bogaert D, Bruin JP, et al. Effect of seven-valent pneumococcal conjugate vaccine on Staphylococcus aureus colonisation in a randomised controlled trial. PLoS ONE. 2011;6:e20229.
Regev-Yochay G, Dagan R, Raz M, Carmeli Y, Shainberg B, Derazne E, et al. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in Children. JAMA. 2004;292:716–20.
Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rümke HC, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363:1871–2.
Shak JR, Vidal JE, Klugman KP. Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol. 2013;21:129-35.
Xu Q, Wischmeyer J, Gonzalez E, Pichichero ME. Nasopharyngeal polymicrobial colonization during health, viral upper respiratory infection and upper respiratory bacterial infection. J Infect. 2017;75:26–34.
Biesbroek G, Wang X, Keijser BJ, Eijkemans RM, Trzcinski K, Rots NY, et al. Seven-valent pneumococcal conjugate vaccine and nasopharyngeal microbiota in healthy children. Emerg Infect Dis. 2014;20:201–10.
Feazel LM, Santorico SA, Robertson CE, Bashraheil M, Scott JA, Frank DN, et al. Effects of vaccination with 10-valent pneumococcal non-typeable Haemophilus influenza [sic] protein d conjugate vaccine (PHiD-CV) on the nasopharyngeal microbiome of Kenyan toddlers. PLoS ONE. 2015;10:e0128064.
Hilty M, Qi W, Brugger SD, Frei L, Agyeman P, Frey PM, et al. Nasopharyngeal microbiota in infants with acute otitis media. J Infect Dis. 2012;205:1048–55.
Mika M, Maurer J, Korten I, Allemann A, Aebi S, Brugger SD, et al. Influence of the pneumococcal conjugate vaccines on the temporal variation of pneumococcal carriage and the nasal microbiota in healthy infants: a longitudinal analysis of a case-control study. Microbiome. 2017;5:85.
Adegbola RA, DeAntonio R, Hill PC, Roca A, Usuf E, Hoet B, et al. Carriage of Streptococcus pneumoniae and other respiratory bacterial pathogens in low and lower-middle income countries: a systematic review and meta-analysis. PLoS ONE. 2014;9:e103293.
Jourdain S, Smeesters PR, Denis O, Dramaix M, Sputael V, Malaviolle X, et al. Differences in nasopharyngeal bacterial carriage in preschool children from different socio-economic origins. Clin Microbiol Infect. 2011;17:907–14.
Russell FM, Carapetis JR, Tikoduadua L, Paeds D, Chandra R, Seduadua A, et al. Invasive pneumococcal disease in Fiji: clinical syndromes, epidemiology, and the potential impact of pneumococcal conjugate vaccine. Pediatr Infec Dis J. 2010;29:870–2.
Dunne EM, Manning J, Russell FM, Robins-Browne RM, Mulholland EK, Satzke C. Effect of pneumococcal vaccination on nasopharyngeal carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus in Fijian children. J Clin Microbiol. 2012;50:1034–8.
Russell FM, Carapetis JR, Satzke C, Tikoduadua L, Waqatakirewa L, Chandra R, et al. Pneumococcal nasopharyngeal carriage following reduced doses of a 7-valent pneumococcal conjugate vaccine and a 23-valent pneumococcal polysaccharide vaccine booster. Clin Vaccine Immunol. 2010;17:1970–6.
Russell FM, Balloch A, Tang ML, Carapetis JR, Licciardi P, Nelson J, et al. Immunogenicity following one, two, or three doses of the 7-valent pneumococcal conjugate vaccine. Vaccine. 2009;27:5685–91.
Russell FM, Licciardi PV, Balloch A, Biaukula V, Tikoduadua L, Carapetis JR, et al. Safety and immunogenicity of the 23-valent pneumococcal polysaccharide vaccine at 12 months of age, following one, two, or three doses of the 7-valent pneumococcal conjugate vaccine in infancy. Vaccine. 2010;28:3086–94.
Russell FM, Carapetis JR, Balloch A, Licciardi PV, Jenney AW, Tikoduadua L, et al. Hyporesponsiveness to re-challenge dose following pneumococcal polysaccharide vaccine at 12 months of age, a randomized controlled trial. Vaccine. 2010;28:3341–9.
Boelsen LK, Dunne EM, Lamb KE, Bright K, Cheung YB, Tikoduadua L, et al. Long-term impact of pneumococcal polysaccharide vaccination on nasopharyngeal carriage in children previously vaccinated with various pneumococcal conjugate vaccine regimes. Vaccine. 2015;33:5708–14.
Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017;8:1162.
Jones EA, Kananurak A, Bevins CL, Hollox EJ, Bakaletz LO. Copy number variation of the beta defensin gene cluster on chromosome 8p influences the bacterial microbiota within the nasopharynx of otitis-prone children. PLoS ONE. 2014;9:e98269.
van Well GT, Sanders MS, Ouburg S, Kumar V, van Furth AM, Morre SA. Single nucleotide polymorphisms in pathogen recognition receptor genes are associated with susceptibility to meningococcal meningitis in a pediatric cohort. PLoS ONE. 2013;8:e64252.
Ioana M, Ferwerda B, Plantinga TS, Stappers M, Oosting M, McCall M, et al. Different patterns of toll-like receptor 2 polymorphisms in populations of various ethnic and geographic origins. Infect Immun. 2012;80:1917–22.
Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191.
Ederveen THA, Ferwerda G, Ahout IM, Vissers M, de Groot R, Boekhorst J, et al. Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome. 2018;6:10.
Schulz JT, Vatucawaqa P, Tuivaga J. 2004 national nutrition survey : main report. Suva: National Food and Nutrition Centre; 2007.
Madan JC, Koestler DC, Stanton BA, Davidson L, Moulton LA, Housman ML, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. MBio. 2012;3.
Hoen AG, Li J, Moulton LA, O’Toole GA, Housman ML, Koestler DC, et al. Associations between gut microbial colonization in early life and respiratory outcomes in cystic fibrosis. J Pediatr. 2015;167:138–47 e1-3.
Neal EFG, Flasche S, Ratu T, Dunne EM, Koyamaibole L, Reyburn RC, et al. Ethnicity and mixing with older children are risk factors for vaccine-type pneumococcal carriage post 10-valent pneumococcal conjugate vaccine introduction in Fiji: a cross-sectional study. Melbourne: 11th International Symposium on Pneumococci and Pneumococcal Diseases; 2018.
Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–52.
Vestrheim DF, Hoiby EA, Aaberge IS, Caugant DA. Impact of a pneumococcal conjugate vaccination program on carriage among children in Norway. Clin Vaccine Immunol. 2010;17:325–34.
Millar EV, O’Brien KL, Watt JP, Bronsdon MA, Dallas J, Whitney CG, et al. Effect of community-wide conjugate pneumococcal vaccine use in infancy on nasopharyngeal carriage through 3 years of age: a cross-sectional study in a high-risk population. Clin Infect Dis. 2006;43:8–15.
Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial communities of the upper respiratory tract and otitis media in children. MBio. 2011;2:e00245–10.
Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–92.
DeMuri GP, Gern JE, Eickhoff JC, Lynch SV, Wald ER. Dynamics of bacterial colonization with Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis during symptomatic and asymptomatic viral upper respiratory tract infection. Clin Infect Dis. 2018;66:1045–53.
Bosch A, de Steenhuijsen Piters WAA, van Houten MA, Chu M, Biesbroek G, Kool J, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. a prospective cohort study. Am J Respir Crit Care Med. 2017;196:1582–90.
Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–15.
de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194:1104–15.
Allen EK, Koeppel AF, Hendley JO, Turner SD, Winther B, Sale MM. Characterization of the nasopharyngeal microbiota in health and during rhinovirus challenge. Microbiome. 2014;2:22.
Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87.
de Goffau MC, Lager S, Salter SJ, Wagner J, Kronbichler A, Charnock-Jones DS, et al. Recognizing the reagent microbiome. Nat Microbiol. 2018;3:851–3.
Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS ONE. 2011;6:e17035.
Jervis-Bardy J, Leong LE, Marri S, Smith RJ, Choo JM, Smith-Vaughan HC, et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome. 2015;3:19.
Stapleton F, Keay LJ, Sanfilippo PG, Katiyar S, Edwards KP, Naduvilath T. Relationship between climate, disease severity, and causative organism for contact lens-associated microbial keratitis in Australia. Am J Ophthalmol. 2007;144:690–8.
Psoter KJ, De Roos AJ, Wakefield J, Mayer J, Rosenfeld M. Season is associated with Pseudomonas aeruginosa acquisition in young children with cystic fibrosis. Clin Microbiol Infect. 2013;19:E483–9.
Kwambana BA, Mohammed NI, Jeffries D, Barer M, Adegbola RA, Antonio M. Differential effects of frozen storage on the molecular detection of bacterial taxa that inhabit the nasopharynx. BMC Clin Pathol. 2011;11(2).
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41.
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6.
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200.
Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: community ecology package. R package version 2.5-1, https://CRAN.R-project.org/package=vegan. 2018.
Calinski T, Harabasz J. A dendrite method for cluster analysis. Commun Stat. 1974;3:1–27.
Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8:e1002687.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–504.
Acknowledgements
The authors wish to thank the study participants and their families, and the staff involved in the Fiji Pneumococcal Project, and the Fiji Ministry of Health and Medical Services. Thank you to Casey Pell and Rosemary Carzino (Murdoch Children's Research Institute) for conducting and/or advising on Pseudomonas culture and identification, and to Kerrie Stevens (Microbiological Diagnostic Unit Public Health Laboratory, The University of Melbourne) for conducting MALDI-TOF MS.
Funding
LB was supported by a Fay Marles Scholarship, The University of Melbourne. CS was supported by an Australian NHMRC Career Development Fellowship and a veski Inspiring Women Fellowship. The Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program. Funders had no role in study design, collection, analysis, and interpretation of data, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
FMR and EKM oversaw and FTR led the field work for the FiPP. CS and MH oversaw the current study. LKB, MM, EMD, SE, FMR, EKM, MH and CS contributed to the study design. LKB and SE conducted laboratory analyses. LKB analysed data with MM overseen by MH and CDN. LKB wrote the manuscript with EMD and CS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
At the time of enrolment in FiPP, consent was given to use swabs for future ethically-approved research. Ethics approval for the work described in this publication was given by Health Sciences Human Ethics Sub-Committee at The University of Melbourne, Australia (Ethics ID #1543881).
Consent for publication
Not applicable
Competing interests
CS, EMD, CDN and EKM are investigators on a clinical research collaboration with Pfizer investigating the impact of PCV on adult pneumonia in Mongolia. MH received consultancy fees and has been awarded investigator initiated research grants from Pfizer, not related to this work. The other authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Participant characteristics. Participant characteristics stratified by ethnicity (Table S1.) and by vaccination status (Table S2.). As well as participant characteristics for those included in this study vs. those not included (Table S3.). (DOCX 22 kb)
Additional file 2:
Richness and Shannon diversity. Plots of richness and Shannon diversity index (Figure S1.) by vaccination status (a and b), by ethnicity (c and d) and by vaccination status within each ethnic group (e and f). (DOCX 674 kb)
Additional file 3:
Species-specific qPCR data. Species-specific qPCR results (carriage prevalence and density) for S. pneumoniae, H. influenzae, M. catarrhalis and S. aureus by vaccination status (Figure S2.), ethnicity (Figure S3.) and vaccination status after stratifying by ethnicity (Figure S4.). (DOCX 954 kb)
Additional file 4:
Association between microbial composition and participant characteristics by PERMANOVA and random forest models. Table (Table S4.) of the PERMANOVA and random forest results for each of the participant characteristics. (DOCX 19 kb)
Additional file 5:
URTI symptoms and ethnicity. Table (Table S5.) of the median relative abundance (%) for the seven most common genera by symptoms of an upper respiratory tract infection (URTI) after stratifying by ethnicity. (DOCX 15 kb)
Additional file 6:
Linear regression data. Unadjusted (Table S6.) and adjusted (Table S7.) linear regression results, as well as this data following log transformation (Tables S8 and S9.) for models that were log transformed. (DOCX 28 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Boelsen, L.K., Dunne, E.M., Mika, M. et al. The association between pneumococcal vaccination, ethnicity, and the nasopharyngeal microbiota of children in Fiji. Microbiome 7, 106 (2019). https://doi.org/10.1186/s40168-019-0716-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40168-019-0716-4