Abstract
Background
The goat (Caprahircus) is one of the most important livestock animals. Goat milk fat is an important component in the nutritional quality of goat milk. Growing evidence points to the critical roles of microRNAs (miRNAs) in lipid metabolism.
Results
Using a highly sensitive method of S-poly(T) plus for miRNAs detection, we analyze the expression patterns of 715 miRNAs in goat mammary gland tissues at different stages of lactation. We observed that miR-25 expression had an inverse relationship with milk production. Overexpression of miR-25 significantly repressed triacylglycerol synthesis and lipid droplet accumulation. To explore the regulatory mechanism of miR-25 in milk lipid metabolism, we analyzed its putative target genes with bioinformatics analysis followed by 3′-UTR assays. Peroxisome proliferative activated receptor gamma coactivator 1 beta (PGC-1beta), a key regulator of lipogenics was identified as a direct target of miR-25 with three specific sites within its 3′-UTR. In addition, miR-25 mimics in goat mammary epithelial cells reduced the expressions of genes involved in lipid metabolism.
Conclusions
Taken together, our results show miR-25 is potentially involved in lipid metabolism and we reveal the function of the miR-25/PGC-1beta regulatory axis during lactation.
Similar content being viewed by others
Background
The goat (Caprahircus) is an important provider of meat and dairy products. Goat milk contains larger amounts of capric, caprylic and medium-chain fatty-acids and smaller globules [1]. These increase the digestibility of goat milk and may promote positive health effects [2]. Analysis of the human consumption of goat and cow milk fat showed that goat milk reduced cholesterol levels but not levels of triglycerides, high-density lipoprotein cholesterol, glutamic oxaloacetic transaminase or glutamic pyruvic transaminase [3]. Thus, goat milk has a higher nutritional value than cow or sheep milk.
Milk fat is a critical component in the nutritional quality of dairy products. The molecular events associated with regulation of milk fat synthesis. For example, lipogenic genes including Acetyl-CoA carboxylase 1 (ACACA), Fatty acid synthase (FASN), stearoyl-CoA desaturase (SCD), Fatty acid desaturase 1 (FADS1), FADS2, 1-acylglycerol-3-phosphate O-acyltransferase 6 (AGPAT6) and glycerol-3-phosphate acyltransferase, mitochondrial (GPAM) are increased until peak-lactation and decrease thereafter [4]. A deeper knowledge of lipid metabolism in the goat mammary gland during lactation is necessary to understand the features of milk, particularly the genes involved in fat metabolism.
MicroRNAs (miRNAs) are non-coding small RNAs that can post-transcriptionally regulate gene expression by pairing with the 3′-untranslated regions (3′-UTRs) or the coding regions of their target mRNAs. The base pairing between miRNA and target gene leads to either degradation of the mRNA or repression of protein translation [5]. Recently, miR-15a, miR-30e and miR-148a have been reported to regulate triacylglycerol synthesis in goat mammary epithelial cells (GMECs) by targeting low-density lipoprotein receptor-related protein 6 (LRP6), yes-associated protein 1 (YAP1) and peroxisome proliferative activated receptor gamma coactivator 1 alpha (PGC-1alpha) [6, 7].
In the present study, we analyzed the miRNA expression patterns of 715 miRNAs using a highly sensitive method of S-poly(T) Plus miRNA real-time PCR [8, 9]. We found that miR-25 is implicated in lipid metabolism during lactation, by directly targeting peroxisome proliferative activated receptor gamma coactivator 1 beta (PGC-1beta), which modulates the expression of sterol regulatory element-binding proteins (SREBPs). Our results establish a miR-25/PGC-1beta regulatory axis in lipid metabolism during lactation.
Methods
Animal tissue samples
Three-year-old Xinong Saanen dairy goats from Northwest A&F University experimental farm were selected and sacrificed for mammary gland tissue collection. All selected goats were of similar body weight and in non-lactation, early lactation (15 d after parturition), peak lactation (60 d after parturition) or late lactation (120 d after parturition) periods. Mammary gland tissues were immediately snap-frozen in liquid nitrogen after washing in diethylpyrocarbonate (DEPC)-treated water. All experimental procedures involving dairy goats were approved by the Institutional Animal Care and Use Committee of the College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi, China.
Cell culture and transfection
Goat mammary epithelial cells (GMECs) were isolated from mammary gland tissue, and purified as previously reported [10]. These cells were treated in a lactogenic medium for 48 h to induce differentiation into secretary cells [11, 12]. For cell experiments, GMECs were cultured in DMEM/F12 medium (Hyclone Laboratories, Beijing, China), containing 5 μg/mL insulin, 5 μg/mL hydrocortisone, 100 U/mL penicillin, 100 μg/mL streptomycin, 10 ng/mL epidermal growth factor 1 (EGF-1, Gibco, Gaithersburg, MD, USA), and 10% fetal bovine serum (FBS, Biological Industries, BeitHaemek, Israel) in a humidified incubator with 5% CO2 at 37 °C. Synthetic miRNA mimics were purchased from RiboBio (Guangzhou, Guangdong, China) and transfected into GMECs using the K2 transfection system (Biontex Laboratories GmbH, München, Germany) according to manufacturer’s instructions.
RNA extraction
Total RNA of tissues and cells was extracted with RNAiso Reagent (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The quality of total RNA was checked by 1% agarose gel electrophoresis. The RNA was quantified using a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) and stored at − 80 °C before use.
Real-time PCR
Mature miRNA expression level was determined using the S-Poly (T) plus method (Geneups, Shenzhen, Guangdong, China).
For miRNA, amplification conditions were as follows: a 10-μL reaction containing 0.2 μg total RNA, 2.5 μL 4× reaction buffer, 1 μL poly A/RT enzyme mix [with 0.8 units of Poly(A) polymerase and 100 units of M-MLV High Performance Reverse Transcriptase] and 1 μL 0.5 μmol/L RT primer. The reaction was performed at 37 °C for 30 min, followed by 42 °C for 30 min, then 75 °C for 5 min. The RT products were amplified and detected using a universal Taqman probe in a 20-μL PCR reaction containing 0.5 μL RT products, 4 μL 5× qPCR probe Mix, 0.5 units Hot Start Polymerase (FAPON, Shenzhen, Guangdong, China), 0.2 mmol/L universal Taqman probe, 0.5 μmol/L forward primer and 0.5 μmol/L universal reverse primer. Primers used were shown in Additional file 1: Table S1. 18S rRNA was used as an internal control. The primers used were as follows: 18S rRNA F: CAGCACATCTTGCGAGTACTC and 18S rRNA R: GTGCAGGGTCCGAGGTCAGAGCCACCTGGGCAATGCAGTGATGGCAAAGG.
For mRNA evaluation, 0.5 μg total RNA was synthesized into cDNA using M-MLV Reverse Transcriptase (TaKaRa) with oligo(dT) 18 plus random hexamer primers (Promega, Madison, Wisconsin, USA). Real-time PCR assays were performed with gene specific primers and SYBR Green PCR Master Mix (Applied Biosystems, Foster, CA, USA). The expression was normalized to ubiquitously expressed transcript ubiquitously expressed transcript protein (UXT).
The PCR reaction was performed at 95 °C for 3 min, followed by 40 amplification cycles consisting of 95 °C for 10 s and 60 °C for 30 s. All real-time PCRs were performed on an ABI StepOneplus real-time PCR System (Applied Biosystems). Primers used for real-time PCR are listed in Additional file 2: Table S2. Relative expression was calculated using the 2 -△△Ct method.
Oil red O staining
Cells were washed three times with phosphate buffered saline (PBS) and then fixed in 10% paraformaldehyde for 1 h at 4 °C. After two washes with PBS, the cells were stained with Oil Red O (0.5 g Oil Red O in 100 mL 70% ethyl alcohol and filtered through a 0.2 μm filter) for 1 h. Cells were then washed thrice with PBS and photographed under a light microscope.
Subsequently, 400 μL of isopropyl alcohol was added to each well, and plates were oscillatedrapidly for about 5 min. Absorbance was then measured at 510 nm. The relative fat droplet content was normalized to the control, and the results of at least three independent experiments were combined.
Triglyceride assay
The amount of intracellular triglyceride relative to total protein was detected using a tissue/cell triacylglycerol assay kit (Applygen Technologies, Beijing, China) and a BCA Protein Assay kit (Thermo Fisher Scientific, Wilmington, DE, USA), respectively.
3′-UTR luciferase reporter assay
We applied TargetScan (http://www.targetscan.org) to predict targets and miRNA binding sites. To generate reporter constructs for luciferase assays, the 3′-UTR of PGC-1beta was PCR amplified from goat genomic DNA and inserted into the pmirGLO dual-luciferase vector (Promega). The primers used were as follows: PGC-1beta WT-1, 5′-CCAGAATTCTCTTCTCCCCATTACACCTTGACCC-3′ (forward) and 5′-CCACTCGAGTCCATTTACCCTGTACCCCTGGACT-3′ (reverse); PGC-1beta WT-2,5′-CCAGAATTCGACTGTATCCACCAGCTACCCAGAT-3′ (forward) and 5′-CCACTCGAGATTCCTCAAGAAACAAAGTTGGGAG-3′ (reverse). To construct mutated 3′-UTR reporter vectors, the predicted miRNA binding sites were mutated by site-directed mutagenesis with the following primers: Muta,5′-CTTTATGTGGGAAGAGAACGTTATAGAAATCTGTCT-3′ (forward) and 5′-TCAGCGAGACAGATTTCTATAACGTTCTCTTCCCACA-3′ (reverse); Mut b,5′-ACCTGGCTAGTGCTTATGACGTTATTGTTTAAGCTGG-3′ (forward) and 5′-TGGGGCCCAGCTTAAACAATAACGTCATAAGCACTAG-3′ (reverse); Mut c,5′-GGAGGGGTTTACTGTAACACGTTATCTGGCAGCCCAG-3′ (forward) and 5′-CAGCAGCTGGGCTGCCAGATAACGTGTTACAGTAAAC-3′ (reverse). All constructs were confirmed by sequencing.
For PGC-1beta 3′-UTR luciferase assays, co-transfection of GMECs with PGC-1beta 3′-UTR or mutated PGC-1beta 3′-UTR reporter plasmids and miR-25 mimics or mimic control was performed with the PEI transfection reagent following the manufacturer’s instructions.
Cells were harvested 48 h after transfection and assayed for Renilla and firefly luciferase activity using the Dual Luciferase Reporter Assay System (Promega) with a luminometer Lumat3 LB9508 (Berthold Technologies, Bad Wildbad, Germany). Firefly luciferase activity was normalized to Renilla luciferase activity.
Western blotting
For western blot analysis, cells were collected and lysed with ice-cold RIPA buffer (50 mmol/L Tris-HCl, pH 7.5; 150 mmol/L NaCl; 1% NP-40; 0.25% sodium deoxycholate, 1 mmol/L EDTA), supplemented with PMSF (Sigma-Aldrich, St. Louis, MO, USA).
Protein concentration was determined using a BCA Protein Assaykit (Thermo Scientific). Proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes (Millipore, USA) and probed with the primary antibodies: polyclonal rabbit anti-PGC-1beta (Cat #22378-1-AP, 1:500 dilution; Proteintech Group) or polyclonal rabbit anti-GAPDH (Cat #HS-PP-0640, 1:1,000 dilution; Hanlin Biotech, Shijiazhuang, China). Polyclonal goat anti-rabbit IgG-HRP (Bio-Rad, Hercules, CA, USA) was used as secondary antibody. The protein bands were visualized using the chemiluminescent ECL western blot detection kit (Advansta, Menlo Park, CA, USA), and images were captured with the Tanon-5200 imaging system (Tanon, Shanghai, China).
Statistical analysis
All results are expressed as the mean ± SD (standard deviation) of at least three triplicates for each treatment. Pairwise comparisons were performed with Student’s t-test using GraphPad Prism 5 software. A P-value of < 0.05 was considered statistically significant.
Results
Differential temporal expression of miRNAs during lactation
To explore potential miRNAs involved in the regulation of lactation, we analyzed the expression patterns of 715 miRNAs from goat mammary glands at four different stages, including non-lactation, early, peak and late lactation using the S-poly(T) plus miRNA quantitative real-time PCR method (Fig. 1a). Among these 715 miRNAs, 122, 143 and 450 are from the domestic goat, sheep and cow databases, respectively (Additional file 3: Table S3). We found that 107 miRNAs were differentially expressed between early and peak lactation (Fig. 1b), and 144 were differentially expressed between peak and late lactation (Fig. 1c). A total of 20 common miRNAs are in the 200 most abundant miRNAs and in the 100 most variable miRNAs across lactation stages (Fig. 1d). Of these 20 miRNAs, levels of miR-17-5p, miR-25, miR-361 and miR-2340 were decreased until peak-lactation and increased thereafter (Fig. 2). Chen et al. revealed that miR-17-5p regulated lipid metabolism during goat lactation [6]. In this study, we focused on miR-25, whose expression level was the highest among these four miRNAs (Additional file 4: Figure S1).
Quantitative real-time PCR analysis of differentially expressed miRNAs in goat mammary glands during lactation. 18S rRNA was used as an internal control. a Workflow of the identification of lactation-related miRNAs. b-c Volcano plot showing the differentially expressed miRNAs at peak lactation compared to early (b) or late (c) lactation. d Heatmap of the miRNAs that were found to be expressed differentially at the four lactation stages. Data for a single pool of three samples at non-lactation and early, peak and late lactation stages are shown. Red to green indicates high to low expression
miR-25 expression is downregulated in goat mammary tissue from non-lactation to late lactation. Real-time PCR validated the expression levels of miR-17-5p, miR-25, miR-361 and miR-2340 in goat mammary tissue at non-lactation, early lactation, peak lactation and late lactation. The data were normalized to 18S. miR-25 expression at the non-lactation stage was set to 1.0. The error bars depict the mean ± S.D. of the samples with three goats at each stage
miR-25 impaired triglyceride and lipid droplet accumulation in GMECs
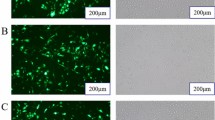
Triacylglycerol droplets and are stored in the cytoplasm as micro lipid droplets [13]. To access the function of miR-25 in lipid metabolism, we analyzed the regulation of triglyceride synthesis and of lipid drop accumulation by miR-25 in goat mammary epithelial cells. We transfected epithelial cells with miR-25 mimics or mimic control (miR-NC). The efficiency of miRNA mimic transfections was confirmed by real-time PCR (Fig. 3a). We found that, miR-25 significantly reduced (0.72-fold, P = 0.0064) the synthesis of triglyceride relative to control (Fig. 3b). Moreover, Oil Red O staining assays showed a reduced number of lipid droplets in miR-25 transfected cells compared to the control (Fig. 3c). Further quantification of lipid droplets confirmed that miR-25 significantly reduced lipid droplet accumulation (0.85-fold, P = 0.0213, Fig. 3d). Taken together, our results show that miR-25 has a repressive role in milk lipid metabolism.
miR-25 represses intracellular triglyceride content and lipid droplet accumulation in GMECs. a Quantitative real-time PCR was performed to measure the expression level of miR-25 in GMECs 48 h after transfection with miR-25 mimics (miR-25), compared with mimic control (miR-NC). miR-17 was used as a positive control. The error bars depict the mean ± S.D. of at least three independent experiments. *P < 0.05. b Chemical analysis showing miR-25 overexpression leading to decreased triglyceride levels in GMECs. Data are shown as the mean ± S.D. of three independent cell samples. **P < 0.01. c Analysis of the presence of lipid droplets in GMECs treated for 48 h with or without miR-25 mimics followed by Oil Red O staining. Scale bar, 200 μm. d Oil Red O-based quantification of fat accumulation in GMECs. Each bar represents the mean ± S.D. for three samples. *P < 0.05
miR-25 repressed the expression of PGC-1beta
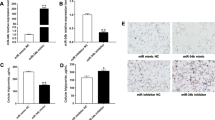
We next explored the mechanism through which miR-25 regulates lipid metabolism by searching for mRNA targets that might mediate its effects in mammary epithelial cells. TargetScan 7.1 (Cambridge, MA, USA) predicted 1,038 genes as its target genes. Further Gene Ontology enrichment analysis using DAVID 6.8 (https://david.ncifcrf.gov) showed that 30 of these genes are lipid metabolism-associated genes (Additional file 5: Table S4). Peroxisome proliferative activated receptor gamma coactivator 1 beta (PGC-1beta) is a well-known regulator of lipogenic genes and is required for the full transcriptional activity of SREBP [13]. To investigate whether PGC-1beta is a potential target gene of miR-25, 48 h after transfection of cells with miR-25 mimic or its control, we analyzed the mRNA and protein levels of PGC-1beta. Due to gradual increases in RNA concentration during lactation, we used UXT as a reference, which is expressed stably across time [14]. We found that miR-25 overexpression resulted in a reduction in the level of PGC-1beta mRNA (0.90-fold, P = 0.0284, Fig. 4a), while western blotting revealed post-transcriptional suppression of PGC-1beta by miR-25 (0.8-fold, P = 0.0155, Fig. 4b). These data demonstrate that PGC-1beta is a probable downstream target of miR-25.
PGC-1beta transcript and protein levels in GMECs transfected with miR-25 mimics or mimic control (miR-NC). a Quantitative real-time PCR was performed to analyze the mRNA levels of PGC-1beta in GMECs using UTX as an internal control. (b, left) Western blot for PGC-1beta in GMECs. (Right) Quantification of PGC-1beta protein levels. miR-25 reduced PGC-1beta protein levels in GMECs. Data represent the mean ± SD of three independent cell samples. **P < 0.01
miR-25 regulates the PGC-1beta gene by directly targeting its 3′-UTR
To understand how miR-25 affects the expression of PGC-1beta, we determined whether there is a direct interaction between miR-25 and PGC-1beta. There are three predicted miR-25 binding sites at the 3′-UTR of PGC-1beta mRNA (sites a, b and c), which are well conserved between goat, sheep, cattle and bison (Fig. 5a). As shown in Fig. 5a, we constructed two wild-type PGC-1beta 3′-UTR luciferase reporter plasmids (WT 1 and 2), and generated their corresponding mutant constructs (Mut a, b, ab and c) with seven mutated residues in the predicted binding sites by site-directed mutagenesis. Enhanced expression of miR-25 significantly repressed the luciferase activities of WT1 and WT2 reporters (0.77-fold, P = 0.0012 and 0.78-fold, P = 0.0017, respectively), whereas these repressions were partly abrogated by Mut a, b or c (Fig. 5b). Moreover, the repression of WT1 was completely abolished when both site a and b were mutated (Fig. 5c). Based on these results, we conclude that miR-25 repressed PGC-1beta expression in goat mammary epithelial cells by directly targeting its 3′-UTR.
miR-25 downregulated PGC-1beta protein expression by directly targeting its 3′-UTR. (a) Schematic illustration of the predicted binding sites of miR-25 in the 3′-UTR of PGC-1beta. Sequence conservation of three predicted miR-25 binding sites within the 3′-UTR of PGC-1beta in four ruminates (Capra hircus, Ovisaries, Bostaurus and bison). WT1 and WT2 represent, respectively, PGC-1beta 3′-UTR-1 (748bp) and PGC-1beta 3′-UTR-2(504bp). The wild type (WT) miRNA-binding sites (black) were modified to complementary sequences (red) to construct the mutated PGC-1beta 3′-UTR (Mut). Mut a, b, c and ab corresponds to mutated PGC-1beta 3′-UTR in site 1, 2, 3 and double mutation of sites 1 and 2, respectively. (b-c) Luciferase reporter assay to assess interaction between miR-25 and PGC-1beta 3′-UTR-1 (b) and PGC-1beta 3′-UTR-2 (c) in GMECs. Data represent the mean ± S.D. of three independent cell samples. *P < 0.05, **P < 0.01, ***P < 0.001
miR-25 modulated other lipid metabolism-related genes in GMECs
Bionaz and Loor [4] identified a regulatory network in lipid metabolism with sterol regulatory element-binding protein 1 (SREBP1), sterol regulatory element-binding protein 2 (SREBP2) and peroxisome proliferator-activated receptor gamma (PPARG) as core regulatory genes. In the SREBP1 subnetwork, PGC-1beta co-activates the SREBP transcription factor and induces the expression of lipogenic genes [13]. Here, we measured the impact of miR-25 overexpression on nine genes in the SREBP1 subnetwork (Fig. 6). miR-25 overexpression had no significant effect on SCD1, ACACA, AGPAT6, insulin induced gene 1 (INSIG1) or PGC-1alpha. However, we found that miR-25 reduced the mRNA levels of SREBP1 (0.86-fold, P = 0.0031), FASN (0.83-fold, P = 0.0368), GPAM (0.92-fold, P = 0.0014) and PPARG (0.75-fold, P = 0.0325).
Regulation of SREBP1, FASN, PPARG, GPAM, ACACA, AGPAT6, SCD1, PGC-1A and ACSL transcript levels in GMECs transfected with miR-25 mimics or mimic control (miR-NC). Cells were collected 48 h after transfection and RNA was isolated and analyzed for mRNA levels of SREBP1 (a), FASN (b), PPARG (c), GPAM (d), ACACA (e), AGPAT6 (f), SCD1 (g), PGC-1A (h) and ACSL (i) by quantitative real-time PCR. For data normalization, UXT was used as an endogenous control. Transcript expression levels are shown as fold change. The data represent the mean ± S.D. of three independent cell samples. * P < 0.05, ** P < 0.01
Discussion
In this study, we analyzed the expression of 715 miRNAs in the goat mammary gland during lactation and characterized miR-25 as a repressor in lipid-metabolism. Our results revealed that miR-25 overexpression leads to repression of PGC-1beta by direct targeting of three different regions within the PGC1-beta 3′-UTR. This reduces the expression of certain lipogenic genes, triglyceride synthesis and lipid droplet accumulation.
During the process of lactation, triacylglycerol accumulation and fatty acid synthesis are essential events that occur in every stage. Twelve miRNAs (miR-15a, miR-17-5p [15], miR-24 [16], miR-26a/b [17], miR-27a [18], miR-30e-5p, miR-130b [19], miR-135a [20], miR-145 [21] and miR-148a [15]) have been shown to repress genes involved in lipid metabolism, including prolactin receptor (PRLR), FASN, LRP6, YAP1, PPARG, INSIG1 and PGC-1alpha during lactation in goat. Evidence suggests that one miRNA may regulate multiple genes, while one gene may be targeted by multiple miRNAs; for example, miR-17 targets PPARA [6], while miR-130b, miR-26a/b and miR-154 target INSIG1. miR-25 joins this miRNA regulatory network as a repressor of lipid synthesis (Fig. 7). Undoubtedly, the functional roles of other unstudied miRNAs remain to be explored.
miR-25 belongs to the miR-25-93-106b cluster, which plays an important role in many malignancies, including ovarian cancer [22], cervicalcancer [23], cholangiocarcinoma [24] and lung cancer [25, 26]. There is growing evidence that miR-25 is involved in lipid metabolism. Liang et al. [27] showed that miR-25 is downregulated during adipocyte differentiation and suppressed 3 T3-L1 adipogenesis by targeting Kruppel-like factor 4 and CCAAT/enhancer-binding protein alpha. Moreover, Hsieh et al. found that miR-25 was downregulated in high-fat diet fed mice relative to low-fat diet fed mice [15]. When viewed together, these findings and our results provide strong evidence to support the involvement of miR-25 in lipid metabolism.
PGC-1beta has long been shown to stimulate the expression of genes involved in lipid metabolism via direct co-activation of the SREBP family [13]. In the present study, decreased SREBP-1c expression by miR-25 overexpression suggests that miR-25 modulates lipid metabolism, at least partially, via the PGC-1beta/SREBP pathway. On the other hand, miR-25 represses target PGC-1beta by binding to multiple sites within its 3′-UTR. Multiple binding sites for the same miRNA can enhance the degree of modulation [14]. In addition, the three miR-25 binding sites within the PGC-1beta 3′-UTR are highly conserved among ruminants, highlighting miR-25 as regulator of PGC-1beta protein expression. These data indicate the importance of miR-25 in regulating PGC-1beta expression.
Conclusion
In conclusion, we revealed miRNA expression patterns in goat mammary gland tissue during lactation and identified miR-25 as lactation related miRNA. We then characterized the role of miR-25 in triglyceride and lipid droplet accumulation and lipid metabolism-related gene expression in GMECs, and determined that miR-25 can repress lipid synthesis via PGC-1beta in GMECs during lactation.
Abbreviations
- ACACA:
-
Acetyl-CoA carboxylase 1
- AGPAT6:
-
1-acylglycerol-3-phosphate O-acyltransferase 6
- FADS1:
-
Fatty acid desaturase 1
- FASN:
-
Fatty acid synthase
- GMEC:
-
Goat mammary epithelial cells
- GPAM:
-
Glycerol-3-phosphate acyltransferase, mitochondrial
- INSIG1:
-
Insulin induced gene 1
- LRP6:
-
Low-density lipoprotein receptor-related protein 6
- PGC-1beta:
-
Peroxisome proliferative activated receptor gamma coactivator 1 beta
- PPARG:
-
Peroxisome proliferator-activated receptor gamma
- PRLR:
-
Prolactin receptor
- SCD:
-
Stearoyl-CoA desaturase
- SREBP1:
-
Sterol regulatory element-binding protein 1
- SREBP2:
-
Sterol regulatory element-binding protein 2
- SREBPs:
-
Sterol regulatory element-binding proteins
- UXT:
-
Ubiquitously expressed transcript protein
- YAP1:
-
Yes-associated protein 1
References
Jandal JM. Comparative aspects of goat and sheep milk. Small Rumin Res. 1996;22:177–85.
Xu M, Wei L, Dai Z, Zhang Y, Li Y, Wang J. Effects of goat milk-based formula on development in weaned rats. Food Nutr Res. 2015;59:28610.
Morita M, Horiuchi M, Kinoshita Y, Yamamoto T, Watanabe T. Relationship between blood triglyceride levels and periodontal status. Community Dent Health. 2004;21:32–6.
Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics. 2008;9:366.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Chen Z, Luo J, Sun S, Cao D, Shi H, Loor JJ. miR-148a and miR-17-5p synergistically regulate milk TAG synthesis via PPARGC1A and PPARA in goat mammary epithelial cells. RNA Biol. 2017;14:326–38.
Chen Z, Qiu H, Ma L, Luo J, Sun S, Kang K, et al. miR-30e-5p and miR-15a synergistically regulate fatty acid metabolism in goat mammary epithelial cells via LRP6 and YAP1. Int J Mol Sci. 2016;17:1909
Niu Y, Zhang L, Qiu H, Wu Y, Wang Z, Zai Y, et al. An improved method for detecting circulating microRNAs with S-poly(T) plus real-time PCR. Sci Rep. 2015;5:15100.
Kang K, Zhang X, Liu H, Wang Z, Zhong J, Huang Z, et al. A novel real-time PCR assay of microRNAs using S-poly(T), a specific oligo(dT) reverse transcription primer with excellent sensitivity and specificity. PLoS One. 2012;7:e48536.
Shi H, Luo J, Zhu J, Li J, Sun Y, Lin X, et al. PPAR gamma regulates genes involved in triacylglycerol synthesis and secretion in mammary gland epithelial cells of dairy goats. PPAR Res. 2013;310948:2013.
Peterson DG, Matitashvili EA, Bauman DE. The inhibitory effect of trans-10, cis-12 CLA on lipid synthesis in bovine mammary epithelial cells involves reduced proteolytic activation of the transcription factor SREBP-1. J Nutr. 2004;134:2523–7.
Kadegowda AKG, Bionaz M, Piperova LS, Erdman RA, Loor JJ. Peroxisome proliferator-activated receptor-γ activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents. J Dairy Sci. 2009;92:4276–89.
Lin J, Yang R, Tarr PT, Wu PH, Handschin C, Li S, et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell. 2005;120:261–73.
Fang Z, Rajewsky N. The impact of miRNA target sites in coding sequences and in 3′UTRs. PLoS One. 2011;6:e18067.
Hsieh CH, Rau CS, Wu SC, Yang JC, Wu YC, Lu TH, et al. Weight-reduction through a low-fat diet causes differential expression of circulating microRNAs in obese C57BL/6 mice. BMC Genomics. 2015;16:699.
Wang H, Luo J, Chen Z, Cao WT, Xu HF, Gou DM, et al. MicroRNA-24 can control triacylglycerol synthesis in goat mammary epithelial cells by targeting the fatty acid synthase gene. J Dairy Sci. 2015;98:9001–14.
Wang H, Luo J, Zhang T, Tian H, Ma Y, Xu H, et al. MicroRNA-26a/b and their host genes synergistically regulate triacylglycerol synthesis by targeting the INSIG1 gene. RNA Biol. 2016;13:500–10.
Tang KQ, Wang YN, Zan LS, Yang WC. miR-27a controls triacylglycerol synthesis in bovine mammary epithelial cells by targeting peroxisome proliferator-activated receptor gamma. J Dairy Sci. 2017;100:4102–12.
Chen Z, Luo J, Ma L, Wang H, Cao W, Xu H, et al. MiR130b-regulation of PPARgamma coactivator- 1alpha suppresses fat metabolism in goat mammary epithelial cells. PLoS One. 2015;10:e0142809.
Ji Z, Dong F, Wang G, Hou L, Liu Z, Chao T, et al. miR-135a targets and regulates prolactin receptor gene in goat mammary epithelial cells. DNA Cell Biol. 2015;34:534–40.
Wang H, Shi H, Luo J, Yi Y, Yao D, Zhang X, et al. miR-145 regulates lipogenesis in goat mammary cells via targeting INSIG1 and epigenetic regulation of lipid-related genes. J Cell Physiol. 2017;232:1030–40.
Zhang H, Zuo Z, Lu X, Wang L, Wang H, Zhu Z. miR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol Rep. 2012;27:594–8.
Song J, Li Y. miR-25-3p reverses epithelial-mesenchymal transition via targeting Sema4C in cisplatin-resistance cervical cancer cells. Cancer Sci. 2017;108:23–31.
Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, et al. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–75.
Xiang J, Hang JB, Che JM, Li HC. miR-25 is up-regulated in non-small cell lung cancer and promotes cell proliferation and motility by targeting FBXW7. Int J Clin Exp Pathol. 2015;8:9147–53.
Wu T, Chen W, Kong D, Li X, Lu H, Liu S, et al. miR-25 targets the modulator of apoptosis 1 gene in lung cancer. Carcinogenesis. 2015;36:925–35.
Liang WC, Wang Y, Liang PP, Pan XQ, Fu WM, Yeung VS, et al. MiR-25 suppresses 3T3-L1 adipogenesis by directly targeting KLF4 and C/EBPalpha. J Cell Biochem. 2015;116:2658–66.
Acknowledgements
We would like to thank Northwest A&F University for providing the goat mammary gland tissues and GMECs. We thank Jeremy Allen, PhD, from LiwenBianji, Edanz Group China, for editing the English text of a draft of this manuscript.
Funding
This work was supported by the Transgenic Project from the Ministry of Agriculture [2014ZX08009-051B to JL], the National Natural Science Foundation of China [81370151 and 81570046 to DG, 31701185 to HQ and 81700054 to YZ]; the Shenzhen Municipal Basic Research Program [JCYJ20150729104027220 to DG and JCYJ20160520174217859 to HQ]; Shenzhen University Interdisciplinary Innovation Team Project [000003 to DG]; Natural Science Foundation of Guangdong Province [2017A030310450 to HQ]; Research Project of Shenzhen Technology University [201731 to HQ].
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Additional files.
Author information
Authors and Affiliations
Contributions
HQ and DG conceived and designed the experiments. LM, ZC and HQ performed the experiments. HQ, JL, LL and DG provided expert advice. All authors analyzed the data. HQ and LM wrote the paper and LL and DG reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The use of animals was in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of China.
Competing interests
The authors declare that they have no competing interests.
Additional files
Additional file 1:
Table S1. Primers used for miRNA real-time PCR. (XLSX 58 kb)
Additional file 2:
Table S2. Primers used for real-time PCR. (DOCX 18 kb)
Additional file 3:
Table S3. List of 715 studied miRNAs. (XLSX 26 kb)
Additional file 4:
Figure S1. Expression levels of miR-17-5p, miR-25, miR-361 and miR-2340 in goat mammary tissue at non-lactation, early lactation, peak lactation and late lactation. Data was presented as Ct value. (DOCX 65 kb)
Additional file 5:
Table S4. 30 lipid metabolism-associated genes. (XLSX 13 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ma, L., Qiu, H., Chen, Z. et al. miR-25 modulates triacylglycerol and lipid accumulation in goat mammary epithelial cells by repressing PGC-1beta. J Animal Sci Biotechnol 9, 48 (2018). https://doi.org/10.1186/s40104-018-0262-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40104-018-0262-0