Abstract
Background
High-altitude pulmonary edema (HAPE) is a life-threatening form of non-cardiogenic edema which occurs in unacclimatized individuals after rapid ascent to high altitude. NR3C1 gene encodes for glucocorticoid receptor (GR) which plays an important role in stress and inflammation. This study aimed to investigate the association of NR3C1 polymorphisms with the susceptibility to HAPE in Han Chinese.
Methods
The 30 SNPs in the NR3C1 gene were genotyped by the Sequenom MassARRAY SNP assay in 133 HAPE patients (HAPE-p) and 135 matched Han Chinese resistant to HAPE (HAPE-r). The genotypic and allele frequencies, odds ratios (ORs), and 95% confidence intervals (95% CIs) were calculated, respectively.
Results
The 12 SNPs showed a significant difference between the HAPE-p and HAPE-r groups. In allelic model analysis, we found that the allele “A” of rs17287745, rs17209237, rs17209251, rs6877893, and rs1866388; the allele “C” of rs6191, rs6188, and rs2918417; the allele “T” of rs33388 and rs4634384; and the allele “G” of rs41423247 and rs10052957 were associated with increased the risk of HAPE. In the genetic model analysis, we found that rs17287745, rs6191, rs6188, rs33388, rs2918417, rs6877893, rs1866388, rs41423247, rs4634384, and rs10052957 were relevant to the increased HAPE risk under the dominant model.
In addition, the haplotype AACACTCAAGTG of the 12 SNPs was detected to be significantly associated with HAPE risk (OR = 2.044, 95%CI = 1.339~3.120, P = 0.0008), while the haplotype GGAGCACGACCG was associated with the decreased risk of HAPE (OR = 0.573, 95% CI = 0.333~0.985, P = 0.0422).
Conclusions
Our findings provide new evidence for the association between SNPs in NR3C1 and an increased risk of HAPE in the Chinese population. NR3C1 polymorphisms are associated with the susceptibility to HAPE in Han Chinese.
Similar content being viewed by others
Background
High-altitude pulmonary edema (HAPE) is non-cardiogenic pulmonary edema that usually occurs at altitudes above 2500 m in rapidly ascending non-acclimatized individuals within the first week after arrival [1, 2]. HAPE is characterized by high pressure in pulmonary arteries, with edema in pulmonary interstitial tissue and alveoli, leading to pulmonary capillary stress failure and a high permeability type of edema. HAPE was defined as a non-inflammatory hemorrhagic pulmonary edema, which may evolve with the features of secondary inflammation [3]. Circulating inflammatory markers of IL-6, IL-1ra, and CRP are upregulated in response to high altitude, and hypoxia-induced inflammation at high altitude may contribute to the development of HAPE [4, 5].
The incidence of HAPE could be reduced by dexamethasone [6]. Glucocorticoids (GCs) have various effects, which are exclusively mediated by the intracellular glucocorticoid receptor (GR). After the binding to GCs, GR is translocated to the nucleus where it interacts with glucocorticoid-responsive elements of different genes to drive gene transcription [7]. In inflammatory reactions, GC inhibits the production of pro-inflammatory cytokines and stimulates the production of anti-inflammatory cytokines [8]. NR3C1 (nuclear receptor subfamily 3, group C, member 1) gene is located on chromosome 5q31-q32 and encodes for the human GR [9]. Variants in the NR3C1 gene may contribute to the spectrum of GC responses in different diseases [7, 10,11,12]. Previously, we screened the variants in exons of the NR3C1 gene and reported that rs6194 polymorphism was correlated with HAPE susceptibility [12]. In this study, we aimed to investigate the association of the polymorphisms in non-coding regions of the NR3C1 gene with the susceptibility to HAPE in Han Chinese.
Materials and methods
Subjects
HAPE patients (HAPE-p) had been hospitalized in Yushu People’s Hospital from 2010 to 2017 owing to the onset of HAPE after arriving 1 to 7 days at Yushu (3760 m) of Qinghai province. The patients were diagnosed with HAPE based on the diagnostic criteria [13]. The blood samples and data were collected. Healthy lowlanders resistant to HAPE (HAPE-r) were enrolled from the same area, with matched age, gender, workplace, and type of work. These subjects had not suffered from HAPE or any other mountain sickness after staying at high altitude for at least 3 months. One hundred thirty-three HAPE and 135 matched HAPE-r were enrolled. This study was approved by the Ethics Committee of the Medical College of Qinghai University, and every subject signed written consent. All subjects were of lowlander Han Chinese ethnicity and had no blood relationship with any other enrolled subject.
Clinical characteristics
Hemoglobin concentration, hematocrit, and percent oxygen saturation were determined from venous blood samples using the Mindray Hematology Analyzer (BC-2300, Shenzhen, People’s Republic of China) and the Pulse Oximeter (Ohmeda 3700 Pulse Oximeter, DatexOhmeda, Boulder, Colorado, USA). Sample collection and DNA extraction were the same with the previous one [12].
SNP selection and genotyping
In this study, a total of 30 SNPs (minor allele frequencies (MAFs) > 5%) in the NR3C1 gene have been identified in the HapMap Han Chinese population and are located in non-coding regions. SNPs were genotyped by the single-base extension detecting technology (iPLEX) (Capital Bio Corporation, Beijing, China). The primers for PCR and single-base extension were designed by using the Sequenom MassARRAY Assay Design Genotyping Software and Tools (Sequenom, San Diego, CA, USA). PCR was performed under the following thermal cycling conditions: 94 °C for 4 min, then 94 °C for 20 s, 56 °C for 30 s, and 72 °C for 1 min for 45 cycles, and 72 °C for 4 min. PCR products were treated with shrimp alkaline phosphatase to remove free deoxyribonucleoside triphosphates, and single-base extension reaction was performed in a system consisted of 2.0 mL of EXTEND MIX, 0.619 mL of ddH2O, 0.94 mL of Extend primer mix, 0.2 mL of iPLEX buffer plus, 0.2 mL of iPLEX terminator, and 0.041 mL of iPLEX enzyme (Sequenom, San Diego, CA, USA). The thermal cycling conditions were as follows: 94 °C for 30 s, then 94 °C for 5 s, 52 °C for 5 s, and 80 °C for 5 s for 40 cycles, and 72 °C for 3 min. The purified extension products were dispensed onto a 384-element SpectroCHIP bioarray (Sequenom, San Diego, CA, USA), and mass spectrometric analysis was performed using the MALDI-TOF (matrix-assisted laser desorption/ionization—time of flight) (Sequenom, San Diego, CA, USA). The results were analyzed using TYPER 4.0 software (Sequenom, San Diego, CA, USA).
Statistical analysis
SPSS software (version 17.0, SPSS, Inc., Chicago, USA) was used for statistical analysis. Haplotype frequencies and the expected number of haplotypes for each individual were performed using SHEsis online software (http://analysis.bio-x.cn). Allele frequencies were calculated based on genotype frequencies in HAPE patients and control subjects, and inter-group differences were estimated by the chi-square test. Deviations from Hardy-Weinberg equilibrium (HWE) were assessed by the chi-square test. The criterion for significance was P < 0.05 for all comparisons.
Results
Basic characteristics of populations
The demographic and clinical characteristics of HAPE-p and HAPE-r are presented in Table 1. We found that percent oxygen saturation was significantly lower whereas heart rate was significantly higher in the HAPE-p group compared to that in the HAPE-r group. As expected, HGB and Hct were higher in the HAPE-r group compared to that in the HAPE-p group. There was no significant difference in age between HAPE-p and HAPE-r groups.
Genotype and allele distribution
We examined the genotypic distributions, allelic frequencies, and associations of 30 SNPs in all subjects. The SNP rs4244032 showed deviations from HWE, while other SNPs were in HWE in both groups (Table 2). The allele and genotype distributions of the 30 SNPs in the HAPE-p and HAPE-r groups are presented in Table 3 (Additional file 1). Twelve SNPs (rs17287745, rs17209237, rs6191, rs17209251, rs6188, rs33388, rs2918417, rs6877893, rs1866388, rs41423247, rs4634384, rs10052957) were significantly associated with HAPE (P < 0.05). In allelic model analysis, we found that the allele “A” of rs17287745, rs17209237, rs17209251, rs6877893, and rs1866388; the allele “C” of rs6191, rs6188, and rs2918417; the allele “T” of rs33388 and rs4634384; and the allele “G” of rs41423247 and rs10052957 were associated with increased the risk of HAPE. In the genetic model analysis, we found that rs17287745, rs6191, rs6188, rs33388, rs2918417, rs6877893, rs1866388, rs41423247, rs4634384, and rs10052957 were relevant to increased HAPE risk under the dominant model.
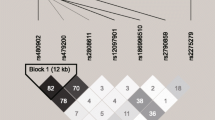
Linkage disequilibrium and haplotype analysis of the 12 SNPs
The locations of these SNPs are shown in Table 2, and the linkage disequilibrium of the SNPs is shown in Fig. 1. In the HAPE-p group, the SNPs were in strong linkage disequilibrium with each other (Fig. 1a). In comparison, few SNPs were found in linkage disequilibrium with each other in the control group (Fig. 1b). The degree of genetic linkage between these SNPs is estimated as D values. Red panels indicate that there exists a strong pairwise linkage disequilibrium between adjacent SNPs; the higher D value is represented with the darker red block, the higher degree of genetic linkage between the SNPs, whereas the white panels indicate that the linkage disequilibrium is weak or non-existent. The haplotype analysis of the 12 SNPs showed a significant difference between the two groups in Table 3, and the haplotype AACACTCAAGTG was detected to be significantly associated with HAPE risk (OR = 2.044, 95%CI = 1.339~3.120, P = 0.0008), while the haplotype GGAGCACGACCG was associated with decreased the risk of HAPE (OR = 0.573, 95% CI = 0.333~0.985, P = 0.0422) (Table 4).
Linkage disequilibrium among the 12 SNPs of the NR3C1 gene in the HAPE-p group (a) and HAPE-r group (b). The degree of genetic linkage between these SNPs is estimated as D values. Red panels indicate that there exists a strong pairwise linkage disequilibrium between adjacent SNPs whereas white panels indicate that the linkage disequilibrium is weak or non-existent
Discussion
It is known that hypoxia can induce inflammation [14]. The levels of circulating proinflammatory markers were increased in healthy volunteers who spent three nights at an elevation higher than 3400 m [4]. In addition, HAPE patients at early stage showed increased counts of alveolar macrophages, neutrophils, and lymphocytes and markedly elevated concentrations of lactate dehydrogenase, IL-1beta, IL-6, IL-8, and TNF-alpha in the bronchoalveolar lavage fluid [15,16,17]. Moreover, vascular leakage, accumulations of inflammatory cells in multiple organs, and elevated serum levels of cytokines were observed in mice exposed to low oxygen concentration [18,19,20]. It has been speculated that hypoxia-induced inflammatory cytokines at high altitude may contribute to the development of HAPE by causing capillary leakage in the lung [4].
Oral administration of dexamethasone is effective in preventing acute mountain sickness (AMS) [6, 18]. Glucocorticoids are regarded as endogenous “dexamethasone” that regulate a broad spectrum of physiologic functions essential to the maintenance of basal and stress-related homeostasis, including inflammatory reactions. The effects of GCs are mediated by GRs, which are steroid/thyroid/retinoic acid nuclear receptor superfamily of transcription factors and function as a ligand-dependent transcription factor that regulates the expression of glucocorticoid-responsive genes. Approximately 20% of the genes expressed in human leukocytes are regulated by GCs [7]. Genetic mutations in NR3C1 have been found to substantially diminish GR function [10]. Therefore, the genetic variations in the NR3C1 gene are important to explain the pathogenesis of diseases.
In this study, we genotyped 30 SNPs of the NR3C1 gene in Han Chinese with and without HAPE. For the first time, we reported significant differences between the two groups in 12 SNPs. Moreover, these polymorphisms were significantly associated with the risk of HAPE. The haplotype [AACACTCAAGTG] may increase the risk of HAPE. This new clue may better explain the genetic variations in the NR3C1 gene contributing to the pathogenesis of HAPE.
Conclusions
Our study suggests that the polymorphisms of NR3C1 gene are associated with the susceptibility to HAPE in Han Chinese.
Change history
23 May 2022
The paper was amended to modify a mistake in one of the supporting funding: (2016-ZJ-7006) was revised to (2019-ZJ-7006).
References
Hackett PH, Roach RC. High-altitude illness. N Engl J Med. 2001;345(2):107.
Basnyat B, Murdoch DR. High-altitude illness. Lancet. 2003;361(9373):1967–74.
Schoene RB, Hackett PH, Henderson WR, et al. High-altitude pulmonary edema. Characteristics of lung lavage fluid. JAMA. 1986;256(1):63–9.
Hartmann G, Tschöp M, Fischer R, et al. High altitude increases circulating interleukin-6, interleukin-1 receptor antagonist and C-reactive protein. Cytokine. 2000;12(3):246–52.
Rosenberger P, Schwab JM, Mirakaj V, et al. Corrigendum: hypoxia-inducible factor-dependent induction of netrin-1 dampens inflammation caused by hypoxia. Nat Immunol. 2009;10(2):195–202.
Maggiorini M, Brunner-La Rocca HP, Peth S, et al. Both tadalafil and dexamethasone may reduce the incidence of high-altitude pulmonary edema: a randomized trial. Ann Intern Med. 2006;145(7):497–506.
Nicolaides NC, Galata Z, Kino T, et al. The human glucocorticoid receptor: molecular basis of biologic function. Steroids. 2010;75(1):1–12.
Neeck G, Renkawitz R, Eggert M. Molecular aspects of glucocorticoid hormone action in rheumatoid arthritis. Cytokines Cell Mol Ther. 2002;7:61–9.
West DV, Eede FVD, Delfavero J, et al. Glucocorticoid receptor gene-based SNP analysis in patients with recurrent major depression. Neuropsychopharmacology. 2006;31(3):620–7.
Zalewski G, Wasilewska A, Zoch-Zwierz W, et al. Response to prednisone in relation to NR3C1 intron B polymorphisms in childhood nephrotic syndrome. Pediatr Nephrol. 2008;23(7):1073–8.
Niu N, Manickam V, Kalari KR, et al. Human glucocorticoid receptor alpha gene (NR3C1) pharmacogenomics: gene resequencing and functional genomics. J Clin Endocrinol Metab. 2009;94(8):3072–84.
Du H, Zhao J, Su Z, Liu Y, Yang Y. Sequencing the exons of human glucocorticoid receptor (NR3C1) gene in Han Chinese with high-altitude pulmonary edema. J Physiol Anthropol. 2018;37(1):7.
Gabry AL, Ledoux X, Mozziconacci M, Martin C. High-altitude pulmonary edema at moderate altitude (< 2,400 m; 7,870 feet) a series of 52 patients. Chest. 2003;123(1):49–53.
Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med. 2011;364(20):1976 author reply 1977.
Grocott M, Martin D, Levett DZ, Mcmorrow R, Windsor J, Montgomery H. Arterial blood gases and oxygen content in climbers on Mount Everest. N Engl J Med. 2009;360(2):140–9.
Thompson LF, Eltzschig HK, Ibla JC, et al. Crucial role for ecto-5’-nucleotidase (CD73) in vascular leakage during hypoxia. J Exp Med. 2004;200(11):1395–405.
Kubo K, Hanaoka M, Hayano T, Miyahara T, Hachiya T, Hayasaka M, Koizumi T, Fujimoto K, Kobayashi T, Honda T. Inflammatory cytokines in BAL fluid and pulmonary hemodynamics in high-altitude pulmonary edema. Respir Physiol. 1998;111(3):301–10.
Zhang C, Qiang Q, Jiang Y, et al. Effects of hypoxia inducible factor-1αon apoptotic inhibition and glucocorticoid receptor downregulation by dexamethasone in AtT-20 cells. BMC Endocr Disord. 2015;15(1):24.
Tang E, Yu C, Luo Y. Dexamethasone for the prevention of acute mountain sickness: systematic review and meta-analysis. Int J Cardiol. 2014;173(2):133–8.
Eltzschig HK, Abdulla P, Hoffman E, et al. HIF-1-dependent repression of equilibrative nucleoside transporter (ENT) in hypoxia. J Exp Med. 2005;202(11):1493.
Acknowledgements
We thanks those physicians for sample collection, Dr Jiangxi from the Yushu People’s Hospital, Dr Nima from the Yushu Bayi Hospital.
Funding
This work were supported by Natural Science Foundation of China (No. 81760335), Qinghai Thousand Talents Program, The 2nd round of “Qinghai 135 high-level talents training project”, Basic Applied Study Foundation of Qinghai (2019-ZJ-7006).
Availability of data and materials
All data and material are freely available.
Author information
Authors and Affiliations
Contributions
YY conceived of the study and drafted the manuscript. HD carried out the molecular genetic studies and performed the statistical analysis. YL, WG, FT, QG, and RLG collected the samples. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Medical College of Qinghai University, and every subject signed written consent.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1. Comparison of genotype distributions and allele frequencies for SNPs associated with HAPE risk under the dominant and recessive model in both groups, respectively. (DOC 306 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yang, Y., Du, H., Li, Y. et al. NR3C1 gene polymorphisms are associated with high-altitude pulmonary edema in Han Chinese. J Physiol Anthropol 38, 4 (2019). https://doi.org/10.1186/s40101-019-0194-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40101-019-0194-1