Abstract
Background
Surgical site infection (SSI) is the most common healthcare-associated infection. We report an outbreak of neurosurgical site infections caused by Serratia marcescens after craniotomy in a tertiary care hospital.
Methods
Between August 6 and 21, 2018, five cases of early-onset SSI caused by S. marcescens after craniotomy were recorded in a 1786-bed tertiary care hospital. Cultures were collected from potential environmental sources and healthcare workers. Whole-genome sequencing (WGS) was used to investigate the genetic relationships among S. marcescens isolates.
Results
The outbreak involved five patients; S. marcescens was isolated from the cerebrospinal fluid, pus, tissue, and blood samples from these patients. S. marcescens was also isolated from shaving razors and brushes. All S. marcescens isolates from the infected patients and razors showed the same resistance patterns on antibiotic-susceptibility tests. WGS revealed close clustering among four of five isolates from the patients and among three of four isolates from the razors. No additional patient developed S. marcescens infection after we stopped using the razors for scalp shaving.
Conclusions
We report an outbreak of neurosurgical site infections after craniotomy, which was associated with shaving razors contaminated by S. marcescens. Shaving scalps with razors should be avoided to prevent SSI.
Similar content being viewed by others
Background
A recent prevalence study found surgical site infections (SSIs) to be the most common healthcare-associated infection (HAI), accounting for 31% of all HAIs among hospitalized patients [1, 2]. SSI, a serious complication of surgery, is associated with significant morbidity, mortality, and health care costs [3, 4]. Serratia marcescens is a gram-negative bacteria widely distributed in the environment. It is a well-known etiologic agent of HAIs, including bacteremia, urinary tract infection, meningitis, pneumonia, and SSIs [5,6,7].
We report an outbreak of neurosurgical site infections by S. marcescens after craniotomy in a tertiary care hospital. We describe the outbreak investigation and the measures taken to control it.
Methods
Background and epidemiological investigation
The outbreak occurred in a 1,786-bed tertiary care hospital in Seoul, South Korea. Between August 6 and 21, 2018, five cases of early-onset SSI caused by S. marcescens after craniotomy were confirmed using the 2018 National Healthcare Safety Network surveillance definitions [8].
The Infection Control Team has performed surveillance of SSIs after craniotomy since 2007 and observed a significant increase in the number of patients with neurosurgical site infection. S. marcescens had hitherto been an uncommon pathogen causing SSI, and the incidence of neurosurgical site infection by S. marcescens significantly increased during the outbreak. As soon as an SSI outbreak was suspected, the team monitored the practices and interviewed personnel in the operating room, neurosurgical ward, and barbershop. Based on information from observation and interviews, environmental samples were obtained for culture. A total of 22 samples were collected from water sources, scrub sinks, surfaces, and shaving razors in the operation room. Another 296 samples were collected from surgical instruments, and 39 nasal samples were collected from doctors, nurses, and other healthcare workers who were associated with neurosurgical operations and the hospital barbershop. An additional 30 samples were collected from the equipment used for preoperative shaving, including razors, brushes, solid soap for shaving, and other tools in the barbershop.
All environmental samples were immersed in 3 mL brain heart infusion (BHI) broth and cultured aerobically for 24 h at 35 °C ± 2 °C. The BHI was inoculated onto blood agar plates and MacConkey agar and incubated for 24 h at 35 °C ± 2 °C. Bacteria were identified using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF), and antimicrobial susceptibility tests were performed using either VITEK 2 or Microscan Walkaway 96 plus instruments.
Molecular biological analysis (microbiological investigation/genotyping)
Whole-genome sequencing (WGS) was performed with nine S. marcescens isolates, including five strains from patients with SSI and four from razors. Genomic DNA extracted using a QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) was quantified using a Qubit®2.0 instrument (Life Technologies, Burlington, Canada), and sequencing libraries were generated using an Illumina TruSeq DNA PCR-Free Kit (15036187; Illumina). Briefly, the samples were normalized to 1 μg DNA and sheared by sonication with a Covaris S220. AMPure XP beads were used for cleanup and size selection, and adapters were then ligated. The fragment sizes for all libraries were measured using a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), and quantitative polymerase chain reaction (qPCR) was performed on a LightCycler® 480 System (Roche, CA, USA) with a KAPA Library Quantification Kit (KK4854, KAPA Biosystems). Whole-genome sequencing was performed on an Illumina Nextseq500 instrument with 300-cycle reagent. To filter adapter and low quality sequences, all reads were trimmed using fastp (version 0.13.1) with default parameters [9]. Kraken2, a taxonomic classification system using exact k-mer matches [10], was used to detect the reference genome and S. marcescens subsp. marcescens Db11 was found to have the closest genome to those of the isolates. The trimmed reads were mapped to the reference sequence using BWA-mem [11], and single nucleotide polymorphisms (SNPs) were detected using SAMtools and bcftools [12] using default parameters except the following: minimum mapping quality score 30, minimum base quality score 20, and ploidy 1. For comparison, using the same process, we processed two S. marcescens genome data, PRJEB28358 and PRJEB27112, publically available in the European Nucleotide Archive (ENA) database consisting of 19 and 17 isolates, derived from the outbreaks in Germany and India, respectively [13, 14]. The core regions were defined as the genomic positions having depths from all 47 isolates ranging between mean depth ± 2 standard deviations. Phylogenetic analysis was conducted using MEGA-X (version 10.0.5) [15] with SNPs only in core regions. The distances between isolates were computed with the maximum composite likelihood method [16], and the phylogenetic tree was built by an unweighted pair group with arithmetic mean (UPGMA) method. The test of phylogeny was performed using the bootstrap test (number of bootstrap = 100).
Statistical analysis
The prevalence of SSI in the pre-outbreak and outbreak periods was compared by chi-square tests using IBM SPSS Statistics for Windows, version 23.0 (IBM Corp., Armonk, NY).
Results
Outbreak investigation
Surveillance of SSIs following craniotomy since 2007 revealed a prevalence of 1.82% (165/9,058) until July 2018. Between August 6 and 21, 2018, five cases (11.63%) out of 43 patients who underwent craniotomy developed neurosurgical SSI due to S. marcescens. The prevalence of neurosurgical site infection increased significantly compared to that in the pre-outbreak period (p = 0.003). Between January 2014 and July 2018, the prevalence of SSI following craniotomy was 1.03% (45/4,384). During this period, the most common etiologic microorganism was Staphylococcus aureus (22.2%), followed by coagulase-negative staphylococcus (21.6%). S. marcescens accounted for 8.3% of etiologic microorganisms. Considering the significant increase in the prevalence of SSI and proportion of S. marcescens, we decided to start an outbreak investigation. Between August 6 and 21, 2018, there was another case of SSI after craniotomy which was attributed to Staphylococcus aureus. Four cases of SSI were monomicrobial infections caused by S. marcescens, and one case was a polymicrobial infection caused by S. marcescens and Klebsiella aerogenes.
The epidemiological and clinical characteristics of the patients are summarized in Table 1. S. marcescens was isolated from cerebrospinal fluid (three patients), pus (four patients), tissue (one patient), and blood (one patient). Three of the five patients (60%) were female, and the mean age of the patients was 39 years (range 11–76 years). The mean interval between surgery and positive culture was 7.2 days (range 3–11 days). These patients were located in different wards and their operations were performed by different surgeons. All patients had undergone preoperative scalp shaving at the barbershop in the study hospital. Among the 387 surveillance samples (22 from the water sink and surface of operating rooms, 296 from surgical instruments, 30 from the barbershop, and 39 from healthcare workers), S. marcescens was isolated from two shaving brushes and four razors that had been used to shave patients’ hair before surgery. Microorganisms other than S. marcescens were detected in 53 samples collected from environment and healthcare workers. (Table 2) All patients were treated with antibiotics, and some underwent wound debridement and/or external ventricular drainage.
Implementation of infection control measures
Upon detection of S. marcescens on the razors, we stopped using the razors for scalp shaving in the barbershop, ward, and operating room. Contamination of operating rooms, instruments, or specific procedures such as wound dressing, and bacterial colonization among health care workers were checked while investigating the outbreak, but there was no common source of S. marcescens infection other than brushes and razors in the barbershop. We recommended that trained doctors or nurses shave scalps using disposable clippers for surgery when shaving was necessary. Infection Control team nurses performed rounds frequently to enforce standardized skin preparation procedures and the use of clippers. After implementation of these infection control measures, no further patients developed S. marcescens infections.
Antibiotic susceptibility and molecular biological analysis
The S. marcescens isolates from infected patients and razors shared the same resistance pattern in antibiotic-susceptibility testing. The isolates were resistant to amoxicillin/clavulanic acid and nitrofurantoin and susceptible to piperacillin/tazobactam, ceftriaxone, cefepime, ertapenem, imipenem, meropenem, gentamicin, ciprofloxacin, and trimethoprim/sulfamethoxazole.
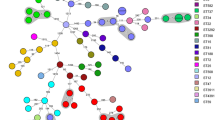
WGS was performed for nine S. marcescens isolates, five strains from patients with SSI, and four from razors. The mean sequencing depth of the nine isolates in this study was 220× (185–244×) and the number of SNPs in the core regions was 142,316 on average. With WGS data of 36 S. marcescens isolates derived from the outbreaks in Germany and India, a total 420,357 bp of core regions with SNPs were used to build the phylogenetic tree. Phylogenetic analysis using WGS revealed close clustering between four of five patient isolates and three of four razor isolates (Fig. 1). S. marcescens isolated from patients and razors were expected to be of the same strain, but there was a considerable distance genetically between the strains isolated from patient 3, razor 1, and others.
The Phylogenetic tree of S. marcescens isolates across three outbreaks in Korea, India, and Germany. An UPGMA phylogenetic tree of S. marcescens isolates from the three outbreaks was generated using the 420,357 core SNP positions detected. The tree reveals the tight clustering of isolates from four patients and three razors in Korea (blue). The other brackets (orange for India, green for Cologne in Germany) annotate clusters described in the previous studies, and all clusters have the same isolates as observed in this study. The small box shows an ~ 100 times enlarged subtree of the clustered isolates. The scale bar and branch lengths are in the units of the number of base substitutions per site. The percentage of replicate trees in which the associated taxa clustered tighter in the bootstrap test (100 replicates) is shown close to the branches in the small box
Discussion
Neurosurgical site infection occurs in 2.2–4.7% of patients after craniotomy [17]. SSI after craniotomy has been associated with increased morbidity, risk of reoperation, neurological sequelae, and mortality. A meta-analysis revealed that the risk factors for SSI included CSF leakage, CSF drainage, operation duration, venous sinus entry, and American Society of Anesthesiologists score [18]. In the current report, we found that contaminated brushes and razors were the sources of the outbreak and that the inadequate shaving technique might cause serious neurosurgical site infections.
It is common practice to shave a patient’s hair around the surgical area. Hair removal has benefits for surgical wound visualization, dressing application, and avoidance of potential annoyance due to hair. Furthermore, many surgeons believe that hair removal is helpful for the prevention of SSI. However, the results of randomized controlled trials revealed no differences in SSIs between patients with and without hair removal before surgery [19]. In the context of neurosurgery, the prevalence of SSIs also does not differ with scalp shaving [20,21,22,23]. Tokimura et al. conducted a retrospective study of 632 patients who underwent cranial surgery without head shaving, of which only 7 (1.1%) developed postoperative infections [20]. Horgan et al. and Ratanalert et al. conducted randomized studies with 20 patients and 225 procedures respectively [22, 23]. In Horgans’ trial, 10 patients had been assigned to the non-shaved group, and there was no infection in both groups. In Ratanalert’s trial, 89 procedures had been allocated to the non-shaved group, and the rates of SSI were 3.37 and 5.88%, respectively. (p > 0.05) Further, a systematic review including 18 studies on the effect of preoperative scalp shaving on SSI showed that preoperative scalp shaving was unnecessary for the asepsis of surgical sites [21]. However, as those studies are underpowered, more research is needed to obtain conclusive evidence. Moreover, the guidelines from the Centers for Diseases Control and Prevention and the World Health Organization recommend that hair not be removed unless it interferes with the surgical procedure [24, 25]. If it is necessary to remove hair, clipping rather than shaving is recommended in light of infection control [19]. In the present study hospital, scalp shaving was performed by barbers in the barbershop. Shaving razors were reused between patients, which eventually resulted in an outbreak of SSI. The infection control team prohibited the use of razors to shave scalps and recommended disposable clippers if scalp shaving was necessary. Following the implementation of these recommendations, no further patients have developed SSI by S. marcescens.
Typing methods for discriminating different bacterial isolates of the same species are essential tools in outbreak investigation. A typing method must have the discriminatory power to distinguish related and unrelated isolates. These methods are classified as phenotypic or molecular. Phenotypic methods include antibiograms, serotyping, and biotyping, while molecular methods include pulse-field gel electrophoresis, multi-locus sequencing typing, and variable number tandem repeat typing. Following improvements in sequencing technologies, whole-genome sequencing (WGS) is a highly discriminative tool that has become the gold standard for bacterial typing, as it can define the complete genomic structure of a pathogen [26, 27]. In this study, we applied WGS to investigate the relatedness of S. marcescens isolates, finding close genetic relationships between four patient and three razor isolates. The strain isolated from patient 3 did not show close genetic similarity with the other strains; therefore the S. marcescens infection in patient 3 should be considered as irrelevant to the S. marcescens outbreak.
We obtained nasal samples rather than samples from the hands for bacterial culture. Considering that bacterial colonizations in the hand sample is a more meaningful indicator of SSI than that in nasal sample, it would be better to consider hands as a site of surveillance culture.
The epidemiological and microbiological evidence indicated that contaminated razors and brushes in the barbershop were the sources of the outbreak. The outbreak ended after stopping preoperative shaving in the barbershop and instead using disposable surgical clippers. Several studies have also reported outbreaks associated with contaminated razors or brushes [28, 29]. The findings of the present report have implications for clinicians and infection preventionists. First, inadequate shaving technique may cause severe SSIs. Second, the use of razors to shave scalps is not recommended because it is associated with an increased incidence of SSI. Third, regular surveillance for SSIs may be beneficial for early outbreak detection. Infection control practitioners can play an important role in screening and preventing outbreaks of SSI based on the prevalence of SSI and the distribution of pathogens.
Conclusions
We reported an outbreak of neurosurgical site infections associated with shaving razors contaminated by S. marcescens. To prevent SSIs, razors should not be used to shave scalps. As recommended in several guidelines, proper hair removal using disposal clippers before surgery is important for the prevention of SSIs.
Availability of data and materials
WGS data were deposited at Short Read Archive (assessment number PRJNA609822). Other data generated or analyzed during this study are included in this published article.
Abbreviations
- SSI:
-
Surgical site infection
- WGS:
-
Whole-genome sequencing
- HAI:
-
Healthcare-associated infection
- BHI:
-
Brain heart infusion
References
Suetens C, Latour K, Kärki T, Ricchizzi E, Kinross P, Moro ML, Jans B, Hopkins S, Hansen S, Lyytikäinen O, et al. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: results from two European point prevalence surveys, 2016 to 2017. Eurosurveillance. 2018;23(46):1800516.
Behnke M, Hansen S, Leistner R, Diaz L, Gropmann A, Sohr D, Gastmeier P, Piening B. Nosocomial infection and antibiotic use a second National Prevalence Study in Germany. Dtsch Arztebl Int. 2013;110:627–33.
de Lissovoy G, Fraeman K, Hutchins V, Murphy D, Song D, Vaughn BB. Surgical site infection: incidence and impact on hospital utilization and treatment costs. Am J Infect Control. 2009;37(5):387–97.
Zimlichman E, Henderson D, Tamir O, Franz C, Song P, Yamin CK, Keohane C, Denham CR, Bates DW. Health care-associated infections: a meta-analysis of costs and financial impact on the US health care system. JAMA Intern Med. 2013;173(22):2039–46.
Us E, Kutlu HH, Tekeli A, Ocal D, Cirpan S, Memikoglu KO. Wound and soft tissue infections of Serratia marcescens in patients receiving wound care: a health care–associated outbreak. Am J Infect Control. 2017;45(4):443–7.
Cilli F, Nazli-Zeka A, Arda B, Sipahi OR, Aksit-Barik S, Kepeli N, Ozinel MA, Gulay Z, Ulusoy S. Serratia marcescens sepsis outbreak caused by contaminated propofol. Am J Infect Control. 2019;47(5):582–4.
Redondo-Bravo L, Gutiérrez-González E, San Juan-Sanz I, Fernández-Jiménez I, Ruiz-Carrascoso G, Gallego-Lombardo S, Sánchez-García L, Elorza-Fernández D, Pellicer-Martínez A, Omeñaca F. Serratia marcescens outbreak in a neonatology unit of a Spanish tertiary hospital: risk factors and control measures. Am J Infect Control. 2019;47(3):271–9.
Centers for Disease Control and Prevention. 2019 Patient Safety Component Manual. http://www.cdc.gov/nhsn/validation. Accessed 20 Sept 2019.
Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90.
Wood DE, Lu J, Langmead B. Improved metagenomic analysis with kraken 2. Genome Biol. 2019;20(1):257.
Yao C, Liu X, Tang Z. Prognostic role of neutrophil-lymphocyte ratio and platelet-lymphocyte ratio for hospital mortality in patients with AECOPD. Int J Chron Obstruct Pulmon Dis. 2017;12:2285–90.
Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–93.
Rohit A, Suresh Kumar D, Dhinakaran I, Joy J, Vijay Kumar D, Kumar Ballamoole K, Karunasagar I, Karola P, Dag H. Whole-genome-based analysis reveals multiclone Serratia marcescens outbreaks in a non-neonatal intensive care unit setting in a tertiary care hospital in India. J Med Microbiol. 2019;68(4):616–21.
Rossen JWA, Dombrecht J, Vanfleteren D, De Bruyne K, van Belkum A, Rosema S, Lokate M, Bathoorn E, Reuter S, Grundmann H, et al. Epidemiological typing of serratia marcescens isolates by whole-genome multilocus sequence typing. J Clin Microbiol. 2019;57(4). https://doi.org/10.1128/JCM.01652-18.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9.
Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101(30):11030–5.
Edwards JR, Peterson KD, Mu Y, Banerjee S, Allen-Bridson K, Morrell G, Dudeck MA, Pollock DA, Horan TC. National Healthcare Safety Network (NHSN) report: data summary for 2006 through 2008, issued December 2009. Am J Infect Control. 2009;37(10):783–805.
Fang C, Zhu T, Zhang P, Xia L, Sun C. Risk factors of neurosurgical site infection after craniotomy: a systematic review and meta-analysis. Am J Infect Control. 2017;45(11):e123–34.
Tanner J, Norrie P, Melen K. Preoperative hair removal to reduce surgical site infection. Cochrane Database Syst Rev. 2011;11.
Tokimura H, Tajitsu K, Tsuchiya M, Yamahata H, Taniguchi A, Takayama K, Kaji M, Hirabaru M, Hirayama T, Shinsato T. Cranial surgery without head shaving. J Cranio-Maxillofac Surg. 2009;37(8):477–80.
Sebastian S. Does preoperative scalp shaving result in fewer postoperative wound infections when compared with no scalp shaving? A systematic review. J Neurosci Nurs. 2012;44(3):149–56.
Horgan MA, Kernan JC, Schwartz MS, Kellogg JX, McMenomey SO, Delashaw JB. Shaveless brain surgery: safe, well tolerated and cost effective. Skull Base Surg. 1999;9(04):253–8.
Ratanalert S, Saehaeng S, Sripairojkul B, Liewchanpattana K, Phuenpathom N. Nonshaved cranial neurosurgery. Surg Neurol. 1999;51(4):458–63.
Mangram AJ, Horan TC, Pearson ML, Silver LC, Jarvis WR, Committee HICPA. Guideline for prevention of surgical site infection, 1999. Infect Control Hosp Epidemiol. 1999;20(4):247–80.
World Health Organization. Global guidelines on the prevention of surgical site infection. https://www.who.int/gpsc/ssi-prevention-guidelines/en/. Accessed 20 Sept 2019.
Kluytmans-van den Bergh MF, Rossen JW, Bruijning-Verhagen PC, Bonten MJ, Friedrich AW, Vandenbroucke-Grauls CM, Willems RJ, Kluytmans JA. Whole-genome multilocus sequence typing of extended-spectrum-beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2016;54(12):2919–27.
Rimoldi SG, Gentile B, Pagani C, Di Gregorio A, Anselmo A, Palozzi AM, Fortunato A, Pittiglio V, Ridolfo AL, Gismondo MR. Whole genome sequencing for the molecular characterization of carbapenem-resistant Klebsiella pneumoniae strains isolated at the Italian ASST Fatebenefratelli Sacco Hospital, 2012–2014. BMC Infect Dis. 2017;17(1):666.
Leng P, Huang W, He T, Wang Y, Zhang H. Outbreak of Serratia marcescens postoperative infection traced to barbers and razors. J Hosp Infect. 2015;89(1):46–50.
Dai Y, Zhang C, Ma X, Chang W, Hu S, Jia H, Huang J, Lu H, Li H, Zhou S. Outbreak of carbapenemase-producing Klebsiella pneumoniae neurosurgical site infections associated with a contaminated shaving razor used for preoperative scalp shaving. Am J Infect Control. 2014;42(7):805–6.
Acknowledgements
We thank our colleagues of infection control team and department of neurosurgery in Seoul National University Hospital.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
EJK and NJK designed the study and wrote the manuscript. WSJ, KIJ and CKK contributed to patients’ medical care. SJK and YRO collected the data. PGC, WBP, EHC and MDO analyzed and interpreted the data. All authors read and approved the final manuscript.
Author’s information
This study was presented at the slide session of surgical site infection in the 5th International Conference on Prevention & Infection Control (ICPIC 2019), 10–13 September 2019, Geneva, Switzerland [abstract no. ICPIC19-ABS-1470].
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of the study hospitals (IRB No. H-1906-146-1043).
Consent for publication
Not applicable.
Competing interests
The authors declare that that have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kim, E.J., Park, W.B., Yoon, JK. et al. Outbreak investigation of Serratia marcescens neurosurgical site infections associated with a contaminated shaving razors. Antimicrob Resist Infect Control 9, 64 (2020). https://doi.org/10.1186/s13756-020-00725-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-020-00725-6