Abstract
Background
The plasmid-encoded multidrug efflux pump oqxAB confers bacterial resistance primarily to olaquindox, quinolones, and chloramphenicol. The aims of this study were to investigate the prevalence of oqxAB among Escherichia coli isolates from dogs, cats, and humans in Henan, China and the susceptibilities of E. coli isolates to common antibiotics.
Methods
From 2012 to 2014, a total of 600 samples which included 400 rectal samples and 200 clinical human specimens were tested for the presence of E. coli. All isolates were screened for oqxAB genes by PCR and sequencing. The MICs of 11 antimicrobial agents were determined by the broth microdilution method. A total of 30 representative oqxAB-positive isolates were subjected to ERIC-PCR and MLST. Additionally, conjugation experiments and southern hybridizations were performed.
Results
Of 270 isolates, 58.5% (62/106) of the isolates from dogs, 56.25% (36/64) of the isolates from cats, and 42.0% (42/100) of the isolates from humans were positive for the oqxAB. Olaquindox resistance was found for 85.7%-100% of oqxAB-positive isolates. Of oqxAB-positive isolates from dogs, cats, and humans, ciprofloxacin resistance was inspected for 85.8%, 59.1%, and 93.8%, respectively. Several oqxAB-positive isolates were demonstrated by ERIC-PCR and MLST, and have high similarity. Phylogenetic analysis showed that oqxAB-positive isolates could be divided into 7 major clusters. OqxAB-positive conjugants were obtained, southern hybridization verified that the oqxAB gene complex was primarily located on plasmids.
Conclusion
In conclusion, oqxAB-positive isolates were widespread in animals and humans in Henan, China. Carriage of oqxAB on plasmids of E. coli isolates may facilitate the emergence of multidrug resistant and its transmission via horizontal transfer, and might pose a potential threat to public health.
Similar content being viewed by others
Background
Nowadays, the growing frequency of antibiotic resistances is a universal problem. Antimicrobial resistance occurs through various mechanisms, such as drug efflux pumps [1, 2]. A novel multidrug efflux pump of Gram-negative bacteria, oqxAB, is a member of the root-nodulation-cell-division (RND) family and was first identified as being encoded by a plasmid-mediated gene that conferred resistance to olaquindox [3]. A number of studies have reported on the occurrence of oqxAB genes primarily in Enterobacteriaceae, including Escherichia coli, Enterobacter cloacae, and Klebsiella pneumoniae isolated from pigs, chickens, humans, and the environment [4–9]. However, the highest positive rates of oqxAB were found in surveys of animals in China [6, 7], and the primary reason might be the widespread use of olaquindox as a production animal growth enhancer [10]. It has been convincingly demonstrated that the genes oqxAB can be horizontally transferred among food-borne pathogens and confer antimicrobial resistance for a variety of antimicrobials, such as quinolones, and chloramphenicol [11, 12]. Because these antimicrobials are a significant part of drug therapy to some human bacterial infections, resistance to these drugs could ultimately pose a significant threat to human health. Thus, investigating prevalence of oqxAB genes in pathogenic E. coli isolates in China is paramount for establishing guidelines for veterinary and human clinical medication use.
Because of the close relationships between humans and their pets, an investigation of multidrug efflux pumps, especially those encoded by the identified oqxAB genes, would be of particular significance for medical science. The aim of this study was to investigate the prevalence of the oqxAB genes among E. coli isolates from companion animals and humans in Henan Province, China.
Methods
Sampling and bacterial isolates
From March 2012 to July 2014, a total of 400 rectal swab samples were recovered from 400 diseased pets (200 dogs and 200 cats) with symptoms of fever, diarrhea and respiratory diseases at three animal hospitals in Henan Agricultural University, ZhengZhou. A total of 200 blood specimens were recovered from adult humans with symptoms of fever, bacteraemia and diarrhea at Henan Provincial People’s Hospital during October 2012 and August 2014. All samples were immediately transported to the laboratory under required preservation conditions (in a cooler with ice) within 6 h of collection, and processed within 2 h for samples to test the presence of E. coli. The samples were incubated in LB media (Beijing Land Bridge Technology Co., Ltd., Beijing, China) at 37 °C overnight for 16-20 h, and draw the line on MacConkey agar plateafter dipping the culture the next day. All presumptive E. coli colonies were identified using VITEK 2 compact automated identification system (BioMérieux, Marcy-I’Etoile, France).
Determination of oqxAB and insertion sequences
A total of 270 strains were screened for oqxA and oqxB genes by PCR using specific primers as described previously [5]. The amplicons obtained were confirmed by sequencing. The whole coding region of the oqxAB gene complex in representative strains isolated from three origins were amplified using primer pairs (producing a fragment of 5140 bp), as reported previously [6]. Then, a PCR product was ligated to a pUC18-T vector (TaKaRa Biotechnology, Dalian, China) and bi-directionally sequenced.
Additionally, the association of insertion sequences ISEcp1 and IS26 with oqxA were also investigated in all oqxAB-positive isolates by PCR using the forward primers ISEcp1-F (5’-GGCCACGTGCATTTTTTATT-3′) and IS26-F (5’-AGCGGTAAATCGTGGAGTGA-3′) and the reverse primer oqxA-R (5’-TCAGGTGAATGTTTCCCCAG-3′) located in oqxA, respectively. The PCR products were sequenced and analyzed with BLAST program to confirm correct amplification.
Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of 11 antimicrobial agents (Additional file 1: Table S1) against the 270 E. coli strains, were determined by the broth microdilution method according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) [13]. E. coli ATCC 25922 was used as a reference strain for quality control in the MIC determinations. The MIC50 and MIC90 were determined which represent concentrations of the relevant antibiotics which inhibited growth of the bacteria by 50% or 90% respectively. The MIC breakpoints for most antimicrobial agents were in accordance with CLSI [13, 14]. The MIC breakpoints for olaquindox and mequindox were based on relevant references [3, 4]. But, if CLSI criteria were not available for some antibiotics, the results were interpreted according to criteria of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [15].
Molecular typing
A total of 30 representative oqxAB-positive E. coli strains (olaquindox-resistant and MIC≥128 μg/mL) selected randomly from humans, dogs and cats were subjected to Enterobacterial Repetitive Intergenic Consensus PCR (ERIC-PCR) using the primers [16]: ERIC-1, 5’-ATGTAAGCTCCTGGGGATTCAC-3′ and ERIC-2, 5’-AAGTAAGTGACTGGGGTGAGCG-3′. Template DNA was extracted using the conventional boiling method, the sample was heated in a thermocycler at 100 °C for 10 min. Immediately, the sample was incubated at − 20 °C for 15 min, and was centrifuged at 12000×g for 15 min. The size of the amplified fragments ranged from 300 to >3000 bp. A similarity coefficient greater than 80% was used to define the same subtype.
Multilocus Sequence Typing (MLST) of these 30 representatives was done by PCR and sequencing. Seven housekeeping genes (aspC, clpX, fadD, icdA, lysP, mdh, and uidA) were amplified and sequenced. The corresponding types (STs) were matched using the electronic database on the E. coli MLST website (http://www.mlst.net/). A phylogenetic tree for the 7 housekeeping gene sequences was constructed using Phylip 3.69 software and their affinity relationships were described.
The phylogenetic groups of the 30 isolates mentioned above were determined by the multiplex PCR-based method, as previously described [17].
Conjugation experiments
Conjugation experiments were done in LB broth or on filters with rifampicin-resistant E. coli C600 as the recipient [18]. Ten oqxAB-positive isolates were randomly selected among 30 oqxAB-positive strains and used as donor strains. Transconjugants were selected on LB agar or MacConkey plates containing olaquindox (64 μg/mL) and rifampicin (360 μg/mL). Antimicrobial susceptibility and detection of transferred oqxAB genes were performed for transconjugants.
Southern hybridization testing
Southern hybridizations were used to test for the plasmid location of oqxAB, according to the procedures as described previously [19]. Plasmid DNA from E. coli was extracted using a Plasmid Midi Kit (QIAGEN, Valencia, CA), according to the manufacturer’s instructions. Plasmid DNA bands were then transferred and cross-linked onto a nylon membrane and were hybridised with digoxigenin (DIG) labeled oqxAB probes using the DIG high prime DNA labeling and detection starter kit (Roche, Mannheim, Germany).
Nucleotide sequencing and submission of oqxAB sequences
The complete nucleotide sequences of the oqxAB genes of E. coli strains Q63, M50, and H050 were submitted to GenBank and were given the accession numbers JX294475, JX412478, and JX469117, respectively.
Data and statistical analysis
The 270 isolated strains were categorized as sensitive (S), resistant (R) based on the MIC values and the CLSI interpretive criteria. For statistical analysis, we carried out chi-square test. A two-sided p-value ≤0.05 were considered to be statistically significant. SPSS 20.0 software (IBM, USA) was used for data analysis.
Results
Isolation and identification of E. coli
A total of 270 E. coli isolates were obtained, which included 106, 64, and 100 E. coli respectively collected from 200 rectal swab samples of dogs, 200 rectal swab samples of cats, and 200 clinical blood samples of humans.
Prevalence of oqxAB
As shown in Table 1, 58.5% (62/106) of the isolates from dogs, 56.25% (36/64) of the isolates from cats, and 42.0% (42/100) of the isolates from humans were positive for the oqxAB. Two common insertion sequences, ISEcp1 and IS26, were investigated in all oqxAB-positive isolates. The whole coding region of the oqxAB genes were amplified and sequenced. ISEcp1, truncated by IS26, was also observed in some oqxAB-positive strains (77/140, 55.0%). Interestingly, most of the oqxAB cassettes of the oqxAB-positive strains (111/140, 79.3%) were also flanked by IS26 similar to composite transposon Tn6010 of K. pneumoniae, which suggested that the dissemination of oqxAB among different E. coli strains might be mediated by the mobile element. This genetic organization (ISEcp1-IS26-oqxAB) is almost identical to corresponding sequences of initially identified plasmid pOLA52 carrying oqxAB genes. The ISEcp1 and IS26 upstream sequences identified here was the first to be associated with the oqxAB genes in E. coli from companion animals in Henan Province, China.
Antimicrobial susceptibility testing
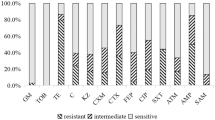
The MICs of 11 common antibiotics against the 270 E. coli isolates from dogs, cats, and humans are shown in Table 1. The MIC50 values of olaquindox for oqxAB-positive strains were 4 to 16-fold higher than those for oqxAB-negative strains (p < 0.01). The MIC50 (1-32 μg/mL) and MIC90 (4-128 μg/mL) values of mequindox were 2 to 8-fold lower than those for olaquindox against both oqxAB-positive and oqxAB-negative isolates. The MIC50 values of ciprofloxacin and florfenicol for oqxAB-positive isolates were all 2 to 16-fold higher than those for oqxAB-negative isolates from the three different origins. Importantly, the MIC50 values of ceftiofur and ceftriaxone were 4-128 μg/mL for both oqxAB-positive and oqxAB-negative isolates, which were much higher than the breakpoints (≤2 μg/mL) for susceptibility. By comparison, the MIC50 values of colistin were 0.125-0.5 μg/mL, which were much lower than the breakpoints (< 2 μg/mL) for susceptibility. Moreover, all isolates from companion animals were susceptible to colistin.
Among oqxAB-positive E. coli strains, resistance rates of ≥68.2% were found for six antibiotics (olaquindox, tetracycline, florfenicol, ceftiofur, doxycycline, and ciprofloxacin). The resistance rates of the oqxAB-positive isolates from dogs and humans to olaquindox were 85.7% and 90.6%. However, the resistance rates to tetracycline, olaquindox, ciprofloxacin, and gatifloxacin were significantly higher among oqxAB-positive isolates than among oqxAB-negative isolates (p < 0.01). Of a total of 270 E. coli strains, more than 59.1% strains were resistant at the same time to ceftiofur and ceftriaxone. However, there were no significant differences in the resistance rates between the oqxAB-positive and oqxAB-negative strains (p > 0.05).
ERIC-PCR, MLST, and phylogenetic analysis
A total of 13 genotypes were identified by ERIC-PCR homology analysis from 30 oqxAB-positive E. coli isolates. Eight isolates (26.7%) primarily belonged to genotype III, followed by genotypes I (7 strains; 23.3%), VI (5 strains; 16.7%), and VIII (4 strains; 13.3%). The remaining each genotypes were found in 1-3 isolates.
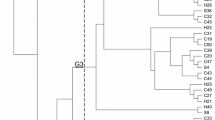
A total of 17 MLST types were identified among these 30 strains from different origins (Fig. 1). Nine novel sequence types (ST3081, ST3082, ST3092, ST3093, ST3094, ST3095, ST3097, ST3098 and ST3099) were detected in this study. The E. coli isolates from dogs, cats, and humans that exhibited identical ERIC-PCR patterns also showed coincident sequence types, for instance, M8 and Q26 (ST48), H029 and Q7 (ST10), Q1 and Q5 (ST155). As shown in Fig. 1, oqxAB-positive isolates could be divided into seven major clusters.
Dendrogram showing the genetic relatedness of 30 E. coli strains (oqxAB-positive). A phylogenetic tree for the seven housekeeping gene sequences was constructed using Phylip 3.69 software and their affinity relationships were described. Notes: Number “M” strains isolated from cats; Number “Q” strains isolated from dogs; Number “H” strains isolated from humans; ND, not determined
Phylogenetic analysis showed that 30 representative strains belonged to group A (23.3%), group B1 (30.0%), group B2 (26.7%), and group D (20.0%), respectively.
Transfer of antimicrobial resistance and the oqxAB genes
Eight transconjugants were successfully obtained from 10 oqxAB-positive isolates by conjugation experiments. Two isolates from dogs did not yield transconjugants. As shown in Table 2, the MICs of mequindox and olaquindox for all transconjugants were similar to those observed for the donors, but were about 8 to 64-fold higher than those observed for the recipients. The oqxAB genes in these 8 oqxAB-positive isolates and mequindox and olaquindox resistance were transferred in the conjugation experiments.
The MICs of florfenicol for the oqxAB transconjugants ranged from 2 to 32 μg/mL, which were about 2 to 32-fold higher than those obtained for the recipients. The MICs of colistin, ceftiofur, and doxycycline for transconjugants (except of J-M50) and donors were all similar and showed distinct increases as compared to recipients.
Localization of the oqxAB genes
Southern hybridization results (Fig. 2b) indicated that oqxAB genes were primarily found in plasmids with sizes of approximately 54 kb, except one strain (Q1 isolated from dog). It was remarkable that the isolates Q63 (isolated from dog) and H050 (isolated from human) yielded two distinct signals located on two plasmids of approximately 54 kb and another unknown size (at least 108 kb), respectively.
Agarose gel electrophoresis (a) and Southern hybridization (b) results for plasmid DNA preparations. Notes: Lanes M1, plasmid Marker V517; 1 to 5, E. coli Q1, M17, Q63, H050, and H029, respectively; M2, Marker 5000. Upper arrow in (b) indicates plasmid DNA; lower arrow in (b) indicates the location of chromosomal DNA and/or sheared plasmid DNA
Discussion
This is the first study to investigate the prevalence and dissemination of oqxAB in E. coli strains isolated from companion animals and from humans in Henan Province, China. It was surprising to find that there was a high prevalence of oqxAB in clinical isolates (51.85%), which was higher than those reported previously in Korea, Denmark, Sweden, Taiwan, and China from food animals and humans (0.06-46.3%) [3–7, 9, 11, 20, 21]. However, the prevalence of oqxAB-positive isolates in this study was lower than previously reported in Iran from urinary tract infections in humans (69.1%) [8]. The major reason may be that olaquindox has been extensively used as a growth promoter for food animals in China.
In agreement with previous studies [5, 11], 79.3% of the oqxAB cassettes of the oqxAB-positive isolates in this study were also flanked by IS26 element similar to Tn6010 of K. pneumoniae, suggesting that these mobile genetic elements are responsible for the dissemination of oqxAB genes [5, 22]. Interestingly, ISEcp1 truncated by IS26 was also observed in 55.0% of oqxAB-positive strains. This genetic organization (ISEcp1-IS26-oqxAB) is almost identical to corresponding sequences of initially identified plasmid pOLA52 carrying oqxAB genes [11]. This suggested that IS26 was inserted in these strains at 3′ end of ISEcp1 as a result of recombinatorial events.
However, this could not account for the significantly high prevalence of oqxAB in clinical isolates from humans, as olaquindox has not been overused for human clinical treatments in China. Probably, oqxAB genes were mostly located on transferable plasmids and could diffuse quickly between human and pet isolates through the food chain [5, 6, 11].
Mequindox, a new synthetic quinoxaline 1,4-dioxide (QdNO) derivative, was developed and widely used in China during the 1990s [10]. It has the same effects as olaquindox as a common animal feed additive to increase the economic benefits of breeding industry. In Table 1, the MIC50 values of mequindox were much lower than those for olaquindox, which differed from a previous report from China [6], and may have been due to its rare use in pets and its limited use in humans.
Excessive antimicrobial use in animals is considered to be the most important contributor to the selection of resistant bacteria [23]. Recently, the use of colistin should be limited in food and companion animals due to reduce the occurrence of MCR-1-harboring strains [24, 25]. Moreover, the use of olaquindox and mequindox also should be limited in food animals due to the high prevalence of the oqxAB gene that we found in China [16]. However, this study demonstrates that oqxAB-positive isolates were not all resistant to olaquindox, which is inconsistent with previous reports [4–7]. This is most probably related to the regulation of oqxAB genes expression. Additionally, it is possible that the oqxAB genes are either poorly expressed or not expressed in these isolates.
The diverse ERIC-PCR patterns among certain oqxAB-positive strains from different origins implied that the horizontal transmission of oqxAB was a possible determinant rather than the direct clonal dissemination between pets and humans, which is consistent with previous reports [26]. A total of 17 MLST types were identified among these 30 oqxAB-positive E. coli isolates from different origins, the same STs of E. coli strains from different origins suggested clonal dissemination of oqxAB-positive strains. Previously, a high prevalence of oqxAB in E. coli isolates associated with predominantly ST238 was reported [27]. However, recent reports showed that the oqxAB-positive E. coli isolates in all belonged to ST533 [28]. In view of this, further studies are needed to investigate the possible transmission of oqxAB genes by either the food chain or by co-infection.
Conjugation experiments and Southern blotting indicated that oqxAB genes were primarily located on plasmids (except of one strain Q1), which is consistent with previous reports [3–7, 11, 29]. However, in this study it was remarkable that there were two hybridization signals found on two plasmids (Fig. 2b) for one isolate from dog and one from human, which indicated they could simultaneously exist on plasmids of different sizes. Meanwhile, oqxAB conferred not only resistance to quinoxalines and chloramphenicol, but also reduced susceptibility to other antimicrobials such as florfenicol and fluoroquinolones. Carriage of oqxAB on plasmids of E. coli isolates may facilitate the emergence of multi-antibiotc resistance and its transmission via horizontal transfer, might pose a potential threat to public health and need for vigilant monitoring these isolates at the human-animal interface.
Conclusion
In conclusion, oqxAB-positive isolates were widespread in pets and humans in Henan Province, China, and the prevalence of oqxAB genes were significantly higher than what was previously found in other countries or areas. Carriage of oqxAB on plasmids of E. coli isolates may facilitate the emergence of multidrug resistant and its transmission via horizontal transfer. The same STs of E. coli strains from different origins suggested clonal dissemination of oqxAB-positive strains. Probably, oqxAB genes were mostly located on transferable plasmids and could diffuse quickly between human and pet isolates through the food chain, this needs to further strengthen supervision and resistance detection. More attention should be paid to the transmission of oqxAB genes in the future.
Abbreviations
- CLSI:
-
Clinical and laboratory standards institute
- EUCAST:
-
European committee on antimicrobial susceptibility testing
- MICs:
-
Minimum inhibitory concentrations
- MLST:
-
Multilocus sequence typing
References
Adewoye L, Topp E, Li XZ. Antimicrobial drug efflux genes and pumps in bacteria of animal and environmental origin. In: Li XZ, Elkins C, Zgurskaya H, editors. Efflux-mediated antimicrobial resistance in bacteria. Adis, Cham: Springer International Publishing; 2016. p. 561–93.
Shafaati M, Boroumand M, Nowroozi J, Amiri P, Kazemian H. Correlation between qacE and qacEΔ1 efflux pump genes, antibiotic and disinfectant resistant among clinical isolates of E. coli. Recent Pa Antiinfec Drug Discov. 2016;11:189–95.
Hansen LH, Johannesen E, Burmølle M, Sørensen AH, Sørensen SJ. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob Agents Chemother. 2004;48:3332–7.
Hansen LH, Sørensen SJ, Jorgensen HS, Jensen LB. The prevalence of the OqxAB multidrug efflux pump amongst olaquindox-resistant Escherichia coli in pigs. Microb Drug Resist. 2005;11:378–82.
Hong BK, Wang M, Chi HP, Kim EC, Jacoby GA, et al. OqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:3582–4.
Zhao JJ, Chen ZL, Chen S, Deng YT, Liu YH, et al. Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farm workers, and the environment. Antimicrob Agents Chemother. 2010;54:4219–24.
Yuan JY, Xu XG, Guo QL, Zhao X, Ye XY, et al. Prevalence of the oqxAB gene complex in Klebsilla pneumoniae and Escherichia coli clinical isolates. J Antimicrob Chemother. 2012;67:1655–9.
Tayebi Z, Heidari H, Kazemian H, Ghafoori SM, Boroumandi S, et al. Comparison of quinolone and beta-lactam resistance among Escherichia coli strains isolated from urinary tract infections. Infez Med. 2016;24:326–30.
Kao CY, Wu HM, Lin WH, Tseng CC, Yan JJ, et al. Plasmid-mediated quinolone resistance determinants in quinolone-resistant Escherichia coli isolated from patients with bacteremia in a university hospital in Taiwan, 2001-2015. Sci Rep. 2016;6:32281.
Huang XJ, Ihsan A, Wang X, Dai MH, Wang YL, et al. Long-term dose-dependent response of Mequindoxon aldosterone, corticosterone and five steroidogenic enzyme mRNAs in the adrenal of male rats. Toxicol Lett. 2009;191:167–73.
Norman A, Hansen LH, She Q, Sørensen SJ. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid. 2008;60:59–74.
Hansen LH, Jensen LB, Sørensen HI, Sørensen SJ. Substrate specificity of the oqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother. 2007;60:145–7.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk and dilution Suseptibility tests for bacteria isolated from animals; approved standard-Fouth edition. CLSI document VET01-A4. Wayne, PA: CLSI; 2013.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-six informational supplement. CLSI document M100-S26. Wayne, PA: CLSI; 2016.
The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 7.0. 2017. [updated 2017.1.1]. Available from: http://www.eucast.org/clinical_breakpoints/.
Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64:159–204.
Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–8.
Chen L, Chen ZL, Liu JH, Zeng ZL, Ma JY, et al. Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J Antimicrob Chemother. 2007;59:880–5.
Liu BT, Wang XM, Liao XP, Sun J, Zhu HQ, et al. Plasmid-mediated quinolone resistance determinants oqxAB and aac(6′)-Ib-cr and extended-spectrum-β-lactamase gene blaCTX-M-24 co-located on the same plasmid in one Escherichia coli strain from China. J Antimicrob Chemother. 2011;66:1638–9.
Ciesielczuk H, Hornsey M, Choi V, Woodford N, Wareham DW. Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J Med Microbiol. 2013;62:1823–7.
Liu BT, Liao XP, Yang SS, Wang XM, Li LL, et al. Detection of mutations in the gyrA and parC genes in Escherichia coli isolates carrying plasmid-mediated quinolone resistance genes from diseased food-producing animals. J Med Microbiol. 2012;61:1591–9.
Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-mediated quinolone resistance. Microbiol Spectr. 2014;2:207–10.
Thomas MG, Smith AJ, Tilyard M. Rising antimicrobial resistance: a strong reason to reduce excessive antimicrobial consumption in New Zealand. New Zeal Med J. 2014;127:72–84.
Huang XH, Yu LF, Chen XJ, Zhi CP, Yao X, et al. High prevalence of Colistin resistance and mcr-1 gene in Escherichia coli isolated from food animals in China. Front Microbiol. 2017;8:562.
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–8.
Macedo NR, Oliveira SR, Lage AP, Santos JL, Araújo MR, et al. ERIC-PCR genotyping of Haemophilus parasuis isolates from Brazilian pigs. Vet J. 2011;188:362–4.
Dotto G, Giacomelli M, Grilli G, Ferrazzi V, Carattoli A, et al. High prevalence of oqxAB in Escherichia coli isolates from domestic and wild lagomorphs in Italy. Microb Drug Resist. 2014;20:118–23.
Chen Y, Sun J, Liao XP, Shao Y, Li L, et al. Impact of enrofloxacin and florfenicol therapy on the spread of oqxAB gene and intestinal microbiota in chickens. Vet Microbiol. 2016;192:1–9.
Fang LX, Li XP, Li L, Li SM, Liao XP, et al. Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci Rep. 2016;6:25312.
Acknowledgements
We thank Ling Yang (South China Agricultural University, CHINA) for reading this manuscript and providing helpful feedback, and key laboratory for animal-derived food safety of Henan Province.
Funding
This study was supported by the National Natural Science Foundation of China (Grant no. 31372481) and the Innovative Support Program for Young Teachers in Universities of Tibet (Grant no. QC2015-77).
Availability of data and materials
The data supporting the findings of this study are included within the manuscript and its supporting information.
Author information
Authors and Affiliations
Contributions
BGL and GZH participated in study conception, design and prepared the manuscript. BGL, YJZ, ZPH, TC, HRS and SMW participated in sample collection and performed the experiments. BGL and HW performed most of the experiments, and reviewed the manuscript. DDH, JHL and YSP analyzed result and reviewed the manuscript. LY and GZH revised the manuscript and coordinated the whole project. All authors read and reviewed the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
With regards to our study’s use of animals, this study protocol was reviewed and approved by the Henan Agriculture University animal ethics committee, and the experiment was performed in accordance with the regulations and guidelines established by this committee. For experiments involving human participants, this study was approved by the Chinese Academy of Sciences human ethics committee, and was carried out strictly in accordance with the approved guidelines. Informed consent was obtained from the hospital.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1: Table S1.
A list of the eleven tested antimicrobials, their classes, their concentrations, and their breakpoints used for susceptibility testing of E. coli. (DOC 58 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, B., Wu, H., Zhai, Y. et al. Prevalence and molecular characterization of oqxAB in clinical Escherichia coli isolates from companion animals and humans in Henan Province, China. Antimicrob Resist Infect Control 7, 18 (2018). https://doi.org/10.1186/s13756-018-0310-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-018-0310-8