Abstract
Background
VIM-type enzyme encodes the most widely acquired metallo-β-lactamases in Gram- negative bacteria. To obtain current epidemiological data for integrons from enterobacteriae in hospital, the study characterizes the genetic structure in In1059 by comparison with In846 integrons harbouring VIM gene and other class 1 integrons including In37, In62, In843 and In1021 with the aim of identifying the putative mechanisms involved integron mobilization and infer evolution of relevant integrons.
Methods
Six of 69 recombinant plasmids from clinical strains were found to be class 1 integrons by digestion with BamHI, drug susceptibility testing, conjugation experiments, PCR amplification, integron cloning and sequencing, genome comparison, and detection of carbapenemase activity.
Results
The sequences of the six recombinant plasmids encoding In1021, In843, In846, In37, In62, and the novel In1059 integron had approximate lengths of ~4.8-, 4.1-, 5.1-, 5.3-, 5.3- and 6.6- kb, respectively. The genetic structures of these integrons were mapped and characterized, and the carbapenemase activities of their parental strains were assessed.
Conclusions
Our results suggest that the six variable integron structures and regular variations that exist in the gene cassettes provide a putative mechanism for the integron changes. Our study has also shown that the genetic features in the integrons named above fall within a scheme involving the stepwise and parallel evolution of class 1 integron variation likely under antibiotic selection pressure in clinical settings.
Similar content being viewed by others
Background
Multidrug resistance can occur by the acquisition of DNA, and the horizontal transfer of drug-resistance genes or drug-resistance gene arrays from one cell to another also occurs and this process transfers drug resistance genes via plasmids and transposons. In recent years, most of the resistance genes found on the plasmids and transposons of Gram-negative bacilli have been found to be integrated into DNA elements called integrons [1]. These integrons contain potential mobile genetic elements that feature a site-specific recombination system capable of integrating and expressing gene cassette structures in the host bacterium [2].
Integrons are the most efficient genetic elements in terms of their ability to disseminate drug-resistance genes between bacteria [3]. Depending on the sequence of the intI gene, integrons have been confirmed to be many classes, and only class 1, 2 and 3 integrons were first identified association with mobile genetic elements [4], intIA with chromosomal integrons and VchintIA with V. cholerae chromosomal integrons [5]. By capturing exogenous cassettes and ensuring expression of the captive genes, an integron plays important role in the horizontal dissemination of antibiotic-resistance genes [6,7,8]. The mechanisms of integration and excision of gene casettes are well described with integrations known to occur at attI × attC recombination sites [9, 10], and excisions requiring attC × attC recombination sites, which occur in single-stranded sequences and activate the folded bottom strand [11, 12]. Because of their linkage with transposons or being plasmid encoded, class 1 integrons can capture genetic structures, express gene cassettes, and facilitate their own mobility, but they are incapable of self-mobilization [5, 13].
Integrons usually contain three functional components: an integrase gene (intI), a primary recombination site (attI), and an outward-orientated promoter (P C ) [14]. Cassette integrations occur mainly at the attI site and this ensures the expression of the captured cassettes under the control of the P C promoter [7, 15]. For most class 1 integrons, the 5′-CS regions are similar and include intI and attI genes; however, differences exist in the 3′-CS sequences and the three open reading frames (ORFs) [16].
VIM-type metallo-β-lactamases were first isolated from Pseudomonas aeruginosa and other Gram-negative nonfermenting bacteria in Europe [17,18,19,20]. These enzymes were subsequently observed worldwide in Gram-negative nonfermentative bacteria and in Enterobacteriaceae. Class 1 integrons harbouring VIM-type gene cassettes have spread among various Gram-negative pathogens [21]. Therefore, this study aimed to characterize the genetic structures of the novel In1059 integron by comparison with the In846 integron and other structural class 1 integrons from enterobacteriaceae in terms of the VIM-type gene cassettes. From this analysis, we have also identified the described mechanism underlying integron mobilization, revealing the relations amongst structures of class 1 integrons including novel In1059 and other integrons in the study.
Methods
Clinical bacterial isolates and drug susceptibility testing
In total, 69 non-redundant multidrug-resistant enterobacteria strains, including non-typhoidal Salmonella Nsa243 and Nsa217 (harbouring In37 and In1021), Enterobacter cloacae Ecl175 (In843), Klebsiella pneumoniae Kpn761 and Kp3349 (In62 and novel In1059), and Enterobacter aerogenes Eae634 (In846), were recovered from hospitalized patients with clinical infections. The strains were assessed for integrons at the Taizhou Municipal Hospital of China (July 2013 to July 2015). Bacterial species was identified by 16 S rRNA gene sequencing [22]. All the above integron-harbouring isolates, plus Escherichia coli TOP10 (Invitrogen, USA), were used in the study. E. coli TOP10 was used as the host for the cloning experiments and for susceptibility testing experiments.
The minimum inhibitory concentration (MIC) values of 12 antimicrobial agents, including cephalosporins (cefazolin, ceftazidime and ceftriaxone), aminoglycosides (netilmicin, tobramycin and amikacin), carbapenems (ertapenem, meropenem and imipenem), and quinolones (norfloxacin, ofloxacin and ciprofloxacin), were determined using drug susceptibility plates and the Microscan broth dilution method (Microscan, Washington, USA). The MICs were interpreted according to the Clinical and Laboratory Standards Institute guidelines [23].
Plasmid digestion, integron cloning and DNA sequencing
Plasmids from six integron-harbouring isolates were collected using an AxyPrep Plasmid Miniprep kit (Axygen Biosciences, Beijing, China) according to the manufacturer’s instructions and according to Wang et al., 2014 [24]. The plasmids were digested with BamHI (TaKaRa, Dalian, China), and the six integron-harbouring recombinant plasmids were electrophoretically resolved to generate genetic maps.
To characterize the recombinant plasmid-encoding integrons from the above six isolates, we digested the plasmids with BamHI and ligated the relevant fragments to the pMD19-T cloning vector (TaKaRa, Dalian, China), and then transformed the ligation mixture into E. coli TOP10 host bacteria. Colonies that were aacA4-positive were isolated and the inserts within the recombinant pMD19-T vectors were sequenced using the primers specified in Table 1 and the following PCR conditions: 3 min at 94 °C, 30 cycles of 1 min each at 94 °C, 50–59 °C and 72 °C, followed by 10 min at 72 °C. The total reaction volume was 25 μL, and the eluent volume was 10 μL. After amplification, the PCR products were separated by gel electrophoresis on a 0.6% agarose gel run at 90 V for 90 min in 0.5 × TBE buffer. Next, the plasmid DNAs or fragments of different sizes that harboured integrons were recovered, and the initial positions of the relevant genes in the recombinant plasmids were determined according to a previously established method for estimating plasmid DNA sizes [25]. After that, the integron sequences involving different genes were obtained, and the genetic structures were mapped and characterized.
Sequence annotation and genome comparison
ORFs were predicted with RAST (http://rast.nmpdr.org/) and further annotated using BLASTP and BLASTN programs (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against UniProtKB/Swiss-Prot (http://web.expasy.org/docs/swiss-prot_guideline.html) and National Center for Biotechnology NR databases (https://www.ncbi.nlm.nih.gov/). Database annotation of drug-resistance genes, mobile elements and other genes was based on CARD (http://arpcard.mcmaster.ca), the β-lactamase (http://www.ncbi.nlm.nih.gov/pathogens/submit_beta_lactamase) database, ISfinder (https://www-is.biotoul.fr/), and INTEGRALL (http://integrall.bio.ua.pt/?). Sequence comparisons were performed with BLASTN and CLUSTALW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Gene organisation diagrams were drawn with Inkscape (https://inkscape. org).
Carbapenemase activity detection
The activities of A, B and D carbapenemase classes in the bacterial cell extracts for the above mentioned six isolates were determined using a modified CarbaNP test [26]. Overnight bacterial cell cultures in Mueller-Hinton (MH) broth were each diluted 1:100 into 3 mL of fresh MH broth, and the bacteria were grown at 37 °C with shaking at 200 rpm to reach OD600s of 1.0 to 1.4. When required, ampicillin was used (200 μg/mL). Bacterial cells were harvested from 2 mL of each culture, and each pellet from the individual cultures was washed twice with 20 mM Tris-HCl (pH 7.8). Each cell pellet was resuspended in 500 μL of 20 mM Tris-HCl (pH 7.8), lysed by sonication and spun at 10000×g at 4 °C for 5 min. Each 50 μL supernatant (containing the enzymatic bacterial suspension fraction) was mixed with 50 μL of the following substrates (I to V), followed by incubation at 37 °C for a maximum of 2 h: substrate I: 0.054% phenol red plus 0.1 mM ZnSO4 (pH 7.8), substrate II: 0.054% phenol red plus 0.1 mM ZnSO4 (pH 7.8), and 0.6 mg/μL imipenem, substrate III: 0.054% phenol red plus 0.1 mM ZnSO4 (pH 7.8), 0.6 mg/μL imipenem, and 0.8 mg/μL tazobactam, substrate IV: 0.054% phenol red plus 0.1 mM ZnSO4 (pH 7.8), 0.6 mg/μL imipenem, and 3 mM EDTA (pH 7.8), and substrate V: 0.054% phenol red plus 0.1 mM ZnSO4 (pH 7.8), 0.6 mg/μL imipenem, 0.8 mg/μL tazobactam, and 3 mM EDTA (pH 7.8).
Nucleotide sequence accession numbers
In1021 Nsa217, In843 Ecl175, In846 Eae634, In37 Nsa243, and In62 Kpn761 sequences, along with In1059 Kp3349, a novel sequence, were deposited in GenBank under the accession numbers KR338349, KR338350, KR338351, KR338352, KJ716225 and KM589496, respectively.
Results and discussion
Integron cloning experiments and antibiotic susceptibility testing
Six of 69 isolates, which resistance to aminoglycoside, quinolone, cephalosporin, and carbapenem antibiotics, from the clinical patients met the requirements of this study. Isolates Kpn761, Eae634, and Kp3349 were collected from blood culture, head wound secretion, and sputum, respectively, from ICU patients. Isolate Ecl175 came from a blood culture from a respiratory medical patient. Isolates Nsa217 and Nsa243 were collected from patient blood cultures in the hospital’s Infectious Disease Unit. Following BamHI digestion and ligation to a pMD19-T cloning vector, the recombinant plasmids were transformed into the competent cells, E. coli TOP10, by heat shock conversion, then, achieved the positive transformants which were selected by blue-white spot experiments, and sequenced. The six integron-containing recombinant plasmids transformed into E. coli TOP10 cells were used for the integron cloning conjugation experiments. The susceptibility test results are listed in Table 2. Concurrently, the six recombinant plasmids were electrophoresed to estimate their sizes (Fig. 1). The susceptibility test results indicated that the conjugation experiments were successful and that the resultant antibiotic resistance was caused by plasmid-mediated genes. The electrophoresis results following BamHI digestion indicated that the sizes of In1021, In843, In846, In37, In62 and the novel In1059 integron were ~4.8 kb, 4.2 kb, 5.1 kb, 5.3 kb, 5.3 kb and 6.6 kb each in length, respectively (Fig. 1). Usually, class 1 integrons that integrate with transposons or are encoded on plasmids can express regular mobilization and transformation capabilities, but they lack self-mobility [5, 13]. In view of these features, integrons can change from one type to another, with the possibility of generating novel types (Fig. 3).
Electrophoretogram of recombinant plasmids encoding In1021, In843, In846, In37, In62, and novel In1059 with genetic mapping. Lane M, λHindIII DNA marker (Amersham Biosciences, Shanghai, China). Lane 1, recombinant plasmids encoding In1021 from the isolate Nsa217 estimated to be approximately 4.8 kb-long; lane 2, recombinant plasmid encoding In843 from the isolate Ecl175 estimated to be approximately 4.1 kb-long; lane 3, recombinant plasmid encoding In846 from the isolate Eae634 estimated to be approximately 5.1 kb-long; lane 4, recombinant plasmid encoding In37 from the isolate Nsa243 estimated to be approximately 5.3 kb-long; lane 5, recombinant plasmid encoding In62 from the isolate Kpn761 estimated to be approximately 5.3 kb-long; lane 6, recombinant plasmid encoding novel In1059 from the isolate Kp3349 estimated to be approximately 6.6 kb-long
Strain carbapenemase activities and genetic features of integrons
The following strains (transformants) In37-TOP10, In846-TOP10 and In1059-TOP10 have class D, B and B carbapenemase activities, respectively, while In37 (isolate Nsa243) appears to have class D activity, while In846 (Eae634) and In1059 (Kp3349) appear to have class B activities, respectively (data not shown). All the above strains were resistant to the cephalosporin, carbapenem and aminoglycoside drugs but they were susceptible to fluoroquinolones (Table 2). In62 apparently represents the most primitive form among these integrons. In62 carries two different resistance markers, while In37, In843, In846, In1021 and In1059 have evolved to capture the determinants of at least three different antibiotic classes, and they most likely confer MDR.
In1059 and In846 integrons differed markedly from the other integrons (In37, In843, In62 and In1021) in this study (Table 3 and Additional file 1). Seemingly, their genetic differences appeared to involve the presence of an additional gene cassette containing bla VIM downstream of the intI1 gene when compared with the other integrons in this study (Fig. 2a). Therefore, the biochemistries of In1059 and In846 should differ from the other integrons. Four gene cassettes and one remnant of dfrA27 were identified in In37 between aacA4cr gene cassette (GCaacA4cr) and GCcatB3, while the existence of GCbla OXA-1 indicates that this integron differs in its biochemistry to In843, In62 and In1021. In37 and In843 differ from In62, In846, In1021 and In1059 class 1 integrons in their 5′-CSs (remnant of intI1), and both lack Pc variant (Table 3 and Additional file 1). In62 appears to have the basic pattern identified in the six class 1 integrons, all of which commonly contain GCaacA4cr. But only GCaacA4cr exists in In62. Notably, one aadA16 and four gene cassetts in the In843 gene are inserted into IS26 in two parts, before reintegrating with the qacE 109 (109 nucleotide remanants of qacE) gene (Table 3 and Additional file 1). Unusually, some fragments such as qacE 109 or qacE 30 combined with GCaadA16 in In843 and In1021, GCarr-3 in In1021 and In846, and GCaadB in In1059 have never been recovered from any class 1 integron to date, and these may have “pseudo-gene cassette” functions (Table 3 and Additional file 1). Indeed, qacE 101 was discovered in class 1 integrons [27], and no qacE 30 or qacE 109 elements have previously been found. In the novel In1059 integron, the GCaacA4 gene coding sequence displays a previously unseen duplication and an unusual 9 bp deletion (cccttcCAT) in its attC, thereby inactivating it (Table 3 and Additional file 1). The duplication extends the sequence to 576-bp in length instead of the usual 555-bp. The resulting gene, which is a novel allele, has been assigned aacA4’-37 (Fig. 3 and Table 3 and Additional file 1), and the fragment itself appears to be embedded within the start of GCaadA16, resulting in a change in its right-hand part of attC site (e.g. the motif locates upstream of the coding sequence), and the remaining part of the attC site in GCaadA16 seems to be a hybrid between the usual one in GCaadA16 and the usual one in GCaacA4 (attC aadA16/aacA4, Table 3 and Additional file 1); furthermore, the GCaadB sequence lacks a right-hand attC part making it look like a pseudo-gene cassette (Table 3 and Additional file 1). However, GCbla VIM-5 appears to be usual attC and normal gene cassette functional in novel In1059 integron.
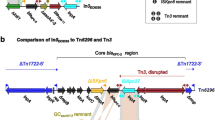
Integron comparisons based on BamHI digestion of plasmids. Genes are denoted by arrows and colours based on the gene function classifications. Shaded areas denote regions with >99% nucleotide sequence identity, with the exception of >97% for the comparison of GCaacA4cr in In846 and GCaacA4′-37 in novel In1059. A, Comparison of In37, In843, In62, In1021, In846 and novel In1059 integrons after BamHI digestion. B-a, Comparison of integrons In37, novel In1059 and transposon Tn1696 (DQ310703); B-b, Comparison of integrons In846, novel In1059 and transposon Tn7017 (KJ571202). The novel In1059 integron shares higher sequence identity with integron Tn7017 than with integron Tn1696
Genetic structures and proposed mobilization and evolution of In0 to In1059 and the analysis of intI1 and attC in class 1 integrons from this study. I, genetic structures and putative mobilization. A, In0; B, A + GCaadB; C, B + GCaadA16; D, C + GCaacA4’-3; E, In1059 = D + GCbla VIM-5 + IS26 + (sul1 + orf5) (replacing sulΔ1); II, Sequences and structures of attI1 (59-be, 59-base elements) and attC. II-a, the attI1 (59-be) site positions are denoted by the bold black line. Open bars indicate potential IntI1 recognition sites, and arrows under the 7-bp core sites indicate relative orientations. The 7-bp core site sequences are shown in bold type, and the strong and weak IntI1-binding sites and direct repeats, DR1 and DR2, are indicated in accordance with the literature [29]. The simple site locations are denoted by an open bar. The “aacaaag(a)” sequences in the black boxes are reminiscent of the spacer in attI1 and occur between the two gene cassettes; recombination or excision of gene cassettes occurs frequently at these sites as indicated in panel I, process C to D. II-b, the attC sequences of class 1 integrons. The bold black letters denote the 7-bp core site, while the numbers are the nucleotide positions (left, negative, right, positive) of the recombination cross-over points, as indicated by a vertical arrow where the recombination or excision of gene cassettes regularly occurs, as shown in panel I. The GenBank accession numbers are shown on the left-hand side of the diagram
Sequence comparisons of InV117 [28] (part of Tn1696, GenBank accession number DQ310703), In1059 and In37 reveal high levels of sequence homology among them (Fig. 2B-a), and In37 in particular shares high sequence identity with In1059. Because the Tn1696 module (including InV117) has the potential to transpose, this suggests that In37 and In1059 should also potentially be mobile elements. Compared with In70 [20] (part of Tn7017, GenBank accession number KJ571202), In1059 and In843 share the most sequence homology with In70 (Fig. 2B-b), and this is especially so for novel In1059. These results provide new insight about the structure of In1059, which coupled with the known plasticity of the In70 genetic context in the recombination plasmids, means that this might have mediated mobilization of the bla VIM-1-containing In70 integron platform [21].
Mobilization and evolutionary inferences for In0 to In1059
Generally, In0 could emerge from excision of all cassettes of an integron. If In0 captures GCaadB on attI1 site likely under antibiotic pressure, structure A might be transferred to structure B, maybe becoming a novel integron (Fig. 3I); then, likely under similar conditions, if joining with GCaadA16, structure B might become structure C, too (Fig. 3I); Also, if joined by GCaacA4’-3, structure C might turn into structure D, perhaps involving a novel integron as well; meanwhile, if inserted into GCbla VIM-5 on attI1 site and IS26 on site between GCaadB and qacE1, then, replaced sul1Δ with sul1 and orf5, structure D will convert into structure E, likely under the right conditions, just as novel In1059 assigned in the study (Fig. 3 and Table 3 and Additional file 1). According to Collis et al., 1998 [29], the recombination or excision of gene cassettes from class 1 integrons frequently occurs at sites of vertical arrow (Fig. 3II-a) between the ″aacaaag(a)″ array and its next nucleotide in attI1, and sites of vertical arrow (Fig. 3II-b) between ″g″ and ″t″ of ″gtttrrry″ array in attC; however, the study appears slightly mutant sequences in terms of the cross-over point of attI1 and attC (Fig. 3II-a and b). Integrons are associated with MDR in Gram-negative bacteria [30, 31, 32] and have been determined as the primary source of drug-resistance genes, and they are also suspected to be reservoirs of drug-resistance genes in patients with bacterial infections [20].
Conclusions
Antibiotic overuse makes it likely that more and more MDR strains will appear in clinical isolates, and most of these strains will contain class 1 integrons. The features of the integrons described above denote a scheme involving the stepwise and parallel evolution of class 1 integron variation likely under antibiotic selection pressure in clinical settings.
References
Hall RM. Gene cassettes and integrons: moving single genes. In: Syvanen M, Kado CI, editors. Horizontal gene transfer. 2nd ed. San Diego: Academic Press; 2002. p. 19–28.
Leverstein-van Hall MA, M Blok HE, T Donders AR, Paauw A, Fluit AC, Verhoef J. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J Infect Dis. 2003;187:251–9.
Huang L-Y, Chen T-L, Lu P-L, Tsai C-A, Cho W-L, Chang F-Y, et al. Dissemination of multidrug-resistant, class 1 integron-carrying Acinetobacter baumannii isolates in Taiwan. Clin Microbiol Infect. 2008;14:1010–9.
Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays inmobile resistance integrons. FEMS Microbiol Rev. 2009;33:757–84.
Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4:608–20.
Hall RM, Stokes HW. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica. 1993;90:115–32.
Collis CM, Hall RM. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–62.
Hall RM, Collis CM. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist Updat. 1998;1:109–19.
MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN. Structural basis for broad DNA-specificity in integron recombination. Nature. 2006;440:1157–62.
Loot C, Ducos-Galand M, Escudero JA, Bouvier M, Mazel D. Replicative resolution of integron cassette insertion. Nucleic Acids Res. 2012;40:8361–70.
Bouvier M, Demarre G, Mazel D. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J. 2005;24:4356–67.
Bouvier M, Ducos-Galand M, Loot C, Bikard D, Mazel D. Structural features of single-stranded integron cassette attC sites and their role in strand selection. PLoS Genet. 2009;5:e1000632.
Carattoli A. Plasmid-mediated antimicrobial resistance in Salmonella Enterica. Curr Issues Mol Biol. 2003;5:113–22.
Collis CM, Kim MJ, Stokes HW, Hall RM. Integron-encoded IntI integrases preferentially recognize the adjacent cognate attI site in recombination with a 59-be site. Mol Microbiol. 2002;46:1415–27.
Levesque C, Brassard S, Lapointe J, Roy PH. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene. 1994;142:49–54.
Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene integration functions: integrons. Mol Microbiol. 1989;3:1669–83.
Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, et al. Cloning and characterization of bla VIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–90.
Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo JD, et al. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid-and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000;44:891–7.
Riccio ML, Pallecchi L, Fontana R, Rossolini GM. In70 of plasmid pAX22, a bla VIM-1-containing integron carrying a new aminoglycoside phosphor-transferase gene cassette. Antimicrob Agents Chemother. 2001;45:1249–53.
Tsakris A, Pournaras S, Woodford N, Palepou MF, Babini GS, Douboyas J, et al. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J Clin Microbiol. 2000;38:1290–2.
Di Pilato V, Pollini S, Rossolini GM. Characterization of plasmid pAX22, encoding VIM-1 metallo-β-lactamase, reveals a new putative mechanism of In70 integron mobilization. J Antimicrob Chemother. 2014;69:67–71.
Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74:2461–70.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, document M100–25. Wayne: Clinical and Laboratory Standards Institute; 2015.
Wang D, Hou W, Chen J, Mou Y, Yang L, Yang L, et al. Characterization of the bla KPC-2 and bla KPC-3 genes and the novel bla KPC-15 gene in Klebsiella pneumoniae. J Med Microbiol. 2014;63:981–7.
Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from shanghai, China. Antimicrob Agents Chemother. 2003;47:2242–8.
Chen Z, Li H, Feng J, Li Y, Chen X, Guo X, et al. NDM-1 encoded by a pNDM- BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front Microbiol. 2015;6:294.
Raherison S, Jove T, Gaschet M, Pinault E, Tabesse A, Torres C, Ploy MC. Expression of the aac(6′)-Ib-cr gene in class 1 integrons. Antimicrob Agents Chemother. 2017;61(5):e02704–16. [Epub ahead of print]
Soler Bistué AJ, Martín FA, Petroni A, Faccone D, Galas M, Tolmasky ME, et al. Vibrio cholerae InV117, a class 1 integron harbouring aac(6′)-Ib and bla CTX-M-2, is linked to transposition genes. Antimicrob Agents Chemother. 2006;50:1903–7.
Collis CM, Kim M-J, Stokes HW, Hall RM. Binding of the purified integron DNA integrase IntI1 to integron and cassette-associated recombination sites. Mol Microbiol. 1998;29:477–90.
Ochman H, Lawrencem JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304.
Sallen B, Rajoharison A, Desvarenne S, Mabilat C. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of Entero- bacteriaceae. Microb Drug Resist. 1995;1:95–202.
Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Brolund A, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009;53:3365–70.
Mazel D, Dychinco B, Webb VA, Davies J. Antibiotic resistance in the EcoR collection: integrons and identification of a novel aad gene. Antimicrob Agents Chemother. 2000;44:1568–74.
Grape M, Farra A, Kronvall G, Sundstrom L. Integrons and gene cassettes in clinical isolates of co-trimoxazole-resistant gram-negative bacteria. Clinic Microbiol Infect. 2005;11:185–92.
Sandvang D, Aarestrup FM, Jensen LB. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett. 1997;157:177–81.
Han HS, Koh YJ, Hur J-S, Jung JS. Occurrence of the strA-strB streptomycin resistance genes in Pseudomonas species isolated from kiwifruit plants. J Microbiol. 2004;42:365–8.
Fihman V, Lartigue MF, Jacquier H, Meunier F, Schnepf N, Raskine L, et al. Appearance of aac(6′)-Ib-cr gene among extended-spectrum β-lactamase- producing Enterobacteriaceae in a French hospital. J Inf Secur. 2008;56:454–9.
Wei Q, Jiang X, Yang Z, Chen N, Chen X, Li G, et al. dfrA27, a new integron- associated trimethoprim resistance gene from Escherichia coli. J Antimicrob Chemother. 2009;63:405–19.
Cámara B, Bielecki P, Kaminski F, dos Santos VM, Plumeier I, Nikodem P, et al. A gene cluster involved in degradation of substituted salicylates via ortho cleavage in Pseudomonas sp. strain MT1 encodes enzymes specifically adapted for transformation of 4-methylcatechol and 3-methylmuconate. J Bacteriol. 2007;189:1664–74.
Levesque C, Piche L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–91.
Acknowledgements
We are grateful to Professor Thomas Jové (INTEGRALL curator), for his designation of In1021, In843, In846, In37, In62 and the novel In1059 integron, all of which are registered in the INTEGRALL database.
Funding
This work was supported by grants from the Foundation of Department of Science and Technology of Zhejiang Province (2014C33153), the Foundation of Zhejiang Health Department (2014KYB218 and 2017KY717) and the Foundation of Taizhou Science and Technology Bureau (14SF05,1301KY36 and 1201KY22).
Availability of data and materials
Please contact author for data requests.
Authors’ contributions
Conceived and designed the experiments: DW. Performed the experiments: all authors. Analyzed the data: DW and DZ. Contributed to the writing of the manuscript: DW. Read and approved the final manuscript: all authors.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The use of patient specimens and all related experimental protocols were approved by the Committee on Human Research at the institutions indicated. The study was carried out in accordance with the approved guidelines of the Ethics Committee of our Hospital, China.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1:
Analysis for integrons: In1059, In843, In846, In37 and In62. (XLS 48 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, D., Yang, J., Fang, M. et al. Characterization of the novel In1059 harbouring VIM gene cassette. Antimicrob Resist Infect Control 6, 50 (2017). https://doi.org/10.1186/s13756-017-0204-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-017-0204-1