Abstract
Background
Vertical integration of the broiler industry allows producers to combine different biosecurity and sanitation practices, housing technologies, and feeding regimens to improve food safety. The purpose of this study was to investigate the prevalence and distribution of Salmonella, to determine the source of Salmonella contaminants, and to reveal the relationships between isolates at each step in the vertically integrated broiler production system in two representative integrated broiler companies in Korea.
Results
A total of 2148 samples were collected from 2 broiler breeder hatcheries, 14 broiler breeder farms, 3 broiler hatcheries, 16 broiler farms, 8 broiler transporting trucks and 6 slaughterhouses belonging to representative integrated broiler companies, and 205 (9.5%) of these samples were positive for Salmonella. The Salmonella prevalence in broiler hatcheries (34.0%) and broiler transporting trucks (62.5%) was higher (P < 0.05) whereas that in the broiler breeder hatchery (0.8%) was lower (P < 0.05), than the overall prevalence. Nine and 13 different Salmonella serotypes were isolated from integrated companies A and B, and the predominant serotypes were S. Virchow (39.7%) and S. Hadar (59.2%), respectively. Pulsed field gel electrophoresis patterns of isolates from the two operations showed significant genetic relatedness within a single system.
Conclusions
In a comparison of the two operations that participated in this study, the prevalence of Salmonella differed significantly between the broiler breeder hatchery, and broiler hatcheries and broiler farms.
Similar content being viewed by others
Background
Salmonella (NTS) are recognized as zoonotic agents and one of the most important foodborne pathogens [23]. Other foodborne pathogens are also considered very important for poultry (e.g Campylobacter). Many foods, particularly poultry and foods originating from poultry, are important sources of food-borne illnesses in humans [13, 25]. Salmonella (23%) is the major cause of bacterial food-borne poisoning in Korea [18]. Cheong et al. [4] reported that poultry are one of the major reservoirs of human Salmonella and Hyeon et al. [10] also reported that chickens and their meat had the highest Salmonella isolation rates among all livestock in Korea.
Dissemination of Salmonella in poultry breeding pyramids have been described previously and is thought to have contributed to the original dissemination of Salmonella to commercial broiler flocks [24, 28]. In general, broiler industry can be divided into the primary breeding sector and the production sector. The production sector range from parent stock to the processing plant and the primary breeding sector from pedigree to grandparent stock. According to a report of the Korea Institute for Animal Products Quality Evaluation [14], a few large integrated companies in Korea own grandparent stock among the primary breeding sector as well as the production sector. In addition, most integrated companies are vertically integrated. Salmonella control in integrated broiler chicken operations is complicated, because there are numerous potential sources of Salmonella contaminants in an integrated poultry system, including chicks, feed, rodents, wild birds, insects, trucks, and the farm and processing plant environments [2].
All sources of Salmonella are potentially important, but it is critical to evaluate the relative importance of different sources in specific management and environmental conditions. The purpose of this study was to investigate the prevalence and distribution of Salmonella, to determine the source of Salmonella contaminants, and to reveal the relationships between isolates at each step in a vertically integrated broiler production system based on two representative integrated broiler companies in Korea.
Methods
Sample collection
A total of 254, 1022, 206, 602, 8 and 128 samples were collected from 2 broiler breeder hatcheries, 14 broiler breeder farms, 3 broiler hatcheries, 16 broiler farms, 8 broiler transport trucks, and 6 slaughterhouses, respectively. All hatcheries, farms, trucks, and slaughterhouses belonged to two representative integrated broiler companies in Korea and were sampled during 10 to 30 visits in each location from 2010 to 2014. Samples were collected from hatcheries by one drag sampling according to a method described by Bailey et al. [2]. Briefly, approximately 300 cm2 in the designated hatcher area was drag swabbed, and each swab placed in a plastic bag. Samples from farms were obtained from dust and feces in accordance with the standard method of the National Poultry Improvement Plan [19]. Briefly, 15 different areas were swabbed per flock with 10 g of dust collected for each sample. Approximately 10 g of feces were also sampled from 15 different locations. Samples from crates and the outside surfaces of transport trucks were collected using the swab method after broiler chickens were unloaded. The rinse water from carcasses in the slaughterhouse were collected according to guidelines of the United States Department of Agriculture, Food Safety and Inspection Service [1]. Four carcasses were sampled from each farm, and each sample was aseptically transferred to a vacuum bag (Cryovag; Sealed Air, USA), 400 ml of sterile buffered peptone water (BPW; Difco, USA) was added, and the bag was shaken 50 times, and approximately 50 ml of rinse water was poured into a sterile specimen cup.
Isolation and identification of Salmonella
Swab samples, feces and 25 ml of rinse water were added to 225 ml of BPW and incubated at 35 ± 2 °C for 20–24 h. After pre-enrichment of the BPW, 0.1 ml of the broth was transferred to a 10 ml of Rappaport-Vassiliadis broth (RV broth; Difco), that was prepared according to the manufacturer’s instructions. The RV broth was incubated overnight at 41.5 °C and streaked onto Rambach agar (Difco). Three typical colonies picked from a plate were serotyped by slide and tube agglutination methods using O and H antisera (Difco) according to the Kauffmann and White scheme [20]. If three colonies from same plate indicated the same serotype, one colony was randomly chosen for inclusion in this study.
Pulsed field gel electrophoresis (PFGE)
PFGE was performed according to the “One-Day (24–28 hr) Standardized Laboratory Protocol for Molecular Subtyping of Non-typhoidal Salmonella by PFGE” [21]. A single colony of each isolate was streaked on tryptic soy agar (TSA) and incubated overnight at 37 °C. Using a cotton swab, a portion of the growth on agar plate was transferred to 2 ml of Cell Suspension Buffer (100 mM Tris: 100 mM EDTA, pH 8.0) and the concentration of cell suspensions adjusted to 14–15% in a bioMerieux Vitek colorimeter. Immediately, 400 μl of adjusted cell suspension was transferred to 1.5 ml micro-centrifuge tubes with 20 μl of proteinase K (20 mg/ml stock), subsequently mixed with 400 μl of melted 1% SeaKem Gold (Cambrex, East Rutherford, NJ): 1% SDS agarose was prepared with TE Buffer (10 mM Tris: 1 mM EDTA, pH 8.0), and pipetted into disposable plug moulds. Three plugs were transferred to 50 ml polypropylene screwtubes with 5 ml of Cell Lysis Buffer (50 mM Tris: 50 mM EDTA, pH 8.0 with sarcosyl) and 25 μl of proteinase K (20 mg/ml stock) and incubated at 54 °C in a shaker water bath for 2 h with agitation. Thereafter, the plugs were washed twice with 15 ml of sterile water and three more times with TE Buffer at 50 °C for 15 min. Chromosomal DNA was digested with 50 U of XbaI (Promega, Madison, WI, USA.), and PFGE was performed on a CHEF Mapper XA System (Bio-Rad Lab., Richmond, CA, USA) in 0.5X Tris-Borate-EDTA buffer (Bio-Rad Lab.) with water circulation at 14 °C. Pulse times were ramped from 2.2 to 63.8 s during an 18 h run at 6.0 V/cm. After electrophoresis, the gels were stained within 2 μg of aqueous ethidium bromide (Sigma-Aldrich. St. Louis, MO, USA) per ml for 15 min and were photographed using 300 nm UV light. Similarities in PFGE patterns were calculated by using a computer-based similarity and clustering program (BioNumerics 3.0, Applied Maths, Biosistematica, Devon, UK). The dice coefficient was used to express similarities, and a similarity matrix was shown graphically by using the unweighted pair group method with arithmetic mean (UPGMA). The relatedness of the PFGE profiles of Salmonella isolates was estimated based on the presence or absence of shared bands. A PFGE type was defined as a group of isolates with similarities ≥85% [12].
Results
Table 1 shows the Salmonella isolates obtained from two representative integrated broiler companies in Korea. A total of 2148 samples were collected from 2 broiler breeder hatcheries, 14 broiler breeder farms, 3 broiler hatcheries, 16 broiler farms, 8 broiler transporting trucks and 6 slaughterhouses belonging to representative integrated broiler companies, 205 (9.5%) of these samples were positive for Salmonella. The Salmonella prevalence in the broiler hatchery (34.0%) and broiler transporting truck (62.5%) was higher (P < 0.05) whereas that in the broiler breeder hatchery (0.8%) was lower (P < 0.05), than the overall prevalence. In integrated broiler company A, Salmonella were most frequently found in the broiler farms (33.3%) and broiler transporting trucks (33.3%), followed by the broiler hatcheries (22.2%), broiler breeder farms (19.0%), carcasses (13.3%), and broiler breeder hatchery (1.6%). In integrated broiler company B, Salmonella were most frequently found in broiler transporting trucks (80.0%), followed by the broiler hatcheries (44.9%), broiler breeder farms (14.7%), broiler farms (13.3%) and carcasses (13.3%), and Salmonella was not isolated from the broiler breeder hatchery. In a comparison of the two companies, the Salmonella prevalence was significantly different between the broiler breeder hatchery, and broiler hatchery and broiler farm (P < 0.05).
Table 2 shows the distribution of Salmonella serotypes recovered from various stages in the two integrated broiler systems. Nine different Salmonella serotypes were isolated from integrated company A, including S. Virchow (39.7%), S. Heidelberg (23.1%), S. Enteritidis (10.3%) and S. Hato (9.0%). Thirteen Salmonella serotypes were isolated from integrated company B, with S. Hadar (59.2%), S. Montevideo (14.5%) and S. Senftenberg (12.8%) the most prevalent types. The distribution of predominant serotypes differed between companies A and B.
Figure 1 shows the distribution of the nine different serotypes of Salmonella isolates in integrated company A. S. Hato was found in the broiler breeder hatchery, broiler breeder farm, broiler hatchery, and chicken slaughterhouse, but was not found in the broiler farms and on the broiler transporting trucks. S. Heidelberg was isolated from 4 out of 5 broiler breeder farms, a broiler hatchery, and a chicken slaughterhouse, and S. Enteritidis was found in a broiler hatchery, 1 out of 6 broiler farms, and a chicken slaughterhouse. S. Virchow was predominant in broiler farms, but was not found in chicken slaughterhouses.
Figure 2 shows the distribution of the 13 different serotypes of Salmonella isolates in integrated company B. Although Salmonella was not found in the broiler breeder hatchery, Salmonella was observed in all subsequent stages of the broiler chicken production and processing system. S. Hadar was isolated from all phases of broiler chicken production and processing including the broiler breeder farms to chicken slaughterhouses. S. Enteritidis was found in 1 out of 2 broiler hatcheries, on the broiler transporting truck and in 3 out of 4 chicken slaughterhouses, but was not found in the 10 broiler farms. S. Typhimurium was only isolated from 1 out of 9 broiler breeder farms.
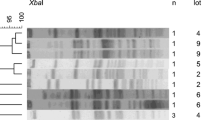
S. Enteritidis (n = 8), S. Montevideo (n = 10) and S. Senftenberg (n = 8), which were isolated frequently in company A and B, were further characterized by PFGE (Fig. 3). PFGE patterns of isolates from the two systems showed significant genetic relatedness within the same system. S. Enteritidis isolated from the hatchery and slaughterhouses of company B clusted together, even when they were isolated one different dates. S. Montevideo showed four PFGE patterns, with isolates from the same origin clustering together.
Discussion
Although Salmonella infection in elite and grandparent chicken breeding flocks is extremely rare and is not considered a source of infection for the industry as a whole, Williams et al. [29] reported that contaminated eggs have the potential spread Salmonella through a hatchery, because eggs from infected flocks can be contaminated during laying, or from infected litter, dust, or equipment at the production site, and subsequently, Salmonella can disseminate throughout the integrated poultry industry. The prevalence of Salmonella (0.8%) in broiler breeder hatcheries in this study was relatively low, but even infected breeder flock can cause widespread Salmonella contamination [27]. Furthermore, the prevalence of Salmonella (15.1%) in broiler breeder farms observed in this study was relatively high compared to that in broiler breeder hatcheries.
The presence and persistence of Salmonella contaminants in commercial hatcheries indicate that the vulnerable day-of-hatch chick may be at greater risk of colonization in the hatchery than during grow-out [6]. Broiler farms usually have a higher prevalence of microorganisms, than other components of the production system because of the relatively poor sanitation management practices on these farms [11]. The prevalence of Salmonella in broiler farms in this study was 15.3%. In other countries, the prevalence of Salmonella in broiler farms range from 9.8% [2] and 13.5% [16] in the United States, to 4.7% and 7.2% in the Netherlands [26], to less than 1% in Finland and Sweden [8], and to as much as 68.2% in Hungary [8].
Previous studies have shown that transport vehicles and crates may play a critical role in transferring Salmonella between broiler farms and slaughterhouses, and that they are important sources of Salmonella contamination for batches of birds and farms [5, 17] because of exposure to the contaminated environments of broiler farms and slaughterhouses. Hald et al. [9] reported that the level of contamination dramatically increases during containment in holding cages before slaughter. In this study, broiler transporting trucks had the highest prevalence of Salmonella (62.5%) of all phases in the broiler supply chain.
In this study, 13.3% of carcasses from chicken slaughterhouses were positive for Salmonella. Lee et al. [15] reported that the prevalence of Salmonella in whole chicken carcasses from slaughterhouses was 15.5%, which was lower than the prevalence (42.7~58.3%) reported in studies conducted before 2011 in Korea [1, 30].
In a comparison of the two operations that participated in this study, the prevalence of Salmonella differed significantly between the broiler breeder hatchery, and broiler hatcheries and broiler farms. Salmonella prevalence might be associated with differences in hygiene and sanitation levels. Vertical integration of the broiler industry allows producers to combine different biosecurity and sanitation practices, housing technologies and feeding regimens to improve food safety. This structure allows a greater ability to govern each aspect of food safety from the breeder farm to the hatchery to the processing plant. Therefore, vertical integration has allowed strict maintenance of biosecurity measures, vaccination programs, and testing for bacteria such as Salmonella at breeder farms and hatcheries.
The predominant serotypes differed between the two operations surveyed. The distribution of Salmonella serotypes may have been affected by inter- and intra-regional differences [2, 3, 7, 22]. The two systems compared in this study were located in the northwest and central areas of Korea. Circulation of Salmonella in these systems may also have been related to farm density, little downtime, and concurrent disease, along with regional density and, poor sanitation and management practices.
PFGE is a useful way to trace Salmonella dissemination within a system. PFGE patterns of S. Enteritidis, S. Montevideo and S. Senftenberg isolated in this study showed significant genetic relatedness within the same system. A number of isolates from farms, hatcheries, and slaughterhouses had PFGE patterns in common, suggesting that many Salmonella strains are transmitted back through stage in the production process. Highly consistent PFGE patterns from different sources and dates within same operation suggest that the propagation of Salmonella clones through the broiler supply chain could spread antibiotic resistance and virulence genes.
Conclusions
In conclusion, this study provides a prevalence and characterization of Salmonella of all phases in the broiler supply chain over the past 5 years in Korea. In a comparison of the two operations that participated in this study, the prevalence of Salmonella differed significantly between the broiler breeder hatchery, and broiler hatcheries and broiler farms. Therefore, these data indicate the critical need to control Salmonella in breeder farms and hatcheries, and suggest important points for control of infection in large-scale poultry operations in Korea.
Abbreviations
- NTS:
-
Non-typhoid Salmonella
- PFGE:
-
Pulsed field gel electrophoresis
- S. Bradford:
-
Salmonella Bradford
- S. Enteritidis:
-
Salmonella Enteritidis
- S. Hadar:
-
Salmonella Hadar
- S. Hato:
-
Salmonella Hato
- S. Havana:
-
Salmonella Havana
- S. Heidelberg:
-
Salmonella Heidelberg
- S. London:
-
Salmonella London
- S. Menston:
-
Salmonella Menston
- S. Montevideo:
-
Salmonella Montevideo
- S. Newport:
-
Salmonella Newport
- S. Reading:
-
Salmonella Reading
- S. Senftenberg:
-
Salmonella Senftenberg
- S. Thompson:
-
Salmonella Thompson
- S. Typhimurium:
-
Salmonella Typhimurium
- S. Ughelli:
-
Salmonella Ughelli
- S. Vellore:
-
Salmonella Vellore
- S. Virchow:
-
Salmonella Virchow
- Salmonella spp.:
-
Salmonella species
- UPGMA:
-
Unweighted pair group method with arithmetic mean
References
Bae DH, Dessie HK, Baek HJ, Kim SK, Lee HS, Lee YJ. Prevalence and characteristics of Salmonella spp. isolated from poultry slaughterhouses in Korea. J Vet Sci. 2013;75:1193–200.
Bailey JS, Stern NJ, Fedorka-Cray P, Craven SE, Cox NA, Cosby DE, Ladely S, Musgrove MT. Musgrove. Sources and movement of Salmonella through integrated poultry operations: a multistate epidemiological investigation. J Food Prot. 2001;11:1690–7.
Byrd JA, DeLoach JR, Corrier DE, Nisbet DJ, Stanker LH. Evaluation of Salmonella serotype distribution from commercial broiler hatcheries and grower houses. Avian Dis. 1999;43:39–47.
Cheong HJ, Lee YJ, Hwang IS, Kee SY, Cheong HW, Song JY, Kim JM, Park YH, Jung JH, Kim WJ. Characteristics of non-typhoidal Salmonella isolates from human and broiler-chickens in southwestern Seoul, Korea. J Korean Med Sci. 2007;22:773–8.
Corry JE, Allen VM, Hudson WR, Breslin MF, Davies RH. Sources of Salmonella on broiler carcasses during transportation and processing: modes of contamination and methods of control. J Appl Microbiol. 2002;92:424–32.
Cox NA, Bailey JS, Blankenship LC, Meinersmann RJ, Stern NJ, McHan F. Fifty percent colonization dose for Salmonella typhimurium administered orally and intracloacally to young broiler chicks. Poult Sci. 1991;69:1809–12.
Cox NA, Bailey JS, Mauldin JM, Blankenship LC, Wilson JL. Extent of salmonellae contamination in breeder hatcheries. Poult Sci. 1991;70:416–8.
European Food Safety Authority. Report of the task force on zoonoses data collection on the analysis of the baseline survey on the prevalence of Salmonella in broiler flocks of Gallus gallus, in the EU, 2005-2006- part a: Salmonella prevalence estimates. EFSA J. 2007;98:1–85.
Hald B, Wedderkopp A, Madsen M. Thermophilic campylobacter spp. in Danish broiler production: a cross-sectional survey and retrospective analysis of risk factors for occurrence in broiler flocks. Avian Pathol. 2000;29:123–31.
Hyeon JY, Chon JW, Hwang IG, Kwak HS, Kim MS, Kim SK, Choi IS, Song CS, Park C, Seo KH. Prevalence, antibiotic resistance, and molecular characterization of Salmonella serovars in retail meat products. J Food Prot. 2011;74:161–6.
Jacobs-Reitsma WF, Bolder NM, Mulder RW. Cecal carriage of Campylobacter and Salmonella in Dutch broiler flocks at slaughter: a one-year study. Poult Sci. 1994;73:1260–6.
Kang ZW, Jung JH, Kim SH, Lee BK, Lee DY, Kim YJ, Lee JY, Won HK, Kim EH, Hahn TW. Genotypic and phenotypic diversity of Salmonella enteritidis isolated from chickens and humans in Korea. J Vet Med Sci. 2009;71:1433–8.
Kim HB, Lee JY, Jang YH, Chang BJ, Kim AR, Choe NH. Prevalence and antimicrobial resistance of Salmonella spp. and Escherichia coli isolated from ducks in Korea. Korean J Vet Res. 2016;56:91–5.
Korea Institute for Animal Products Quality Evaluation. Research on the actual conditions of livestock distribution system. 2014. http://www.ekape.or.kr/view/user/main/main.asp. Accessed Nov 2014.
Lee SK, Choi D, Kim HS, Kim DH, Seo KH. Prevalence, seasonal occurrence, and antimicrobial resistance of Salmonella spp. isolates recovered from chicken carcasses sampled at major poultry processing plants of South Korea. Foodborne Pathog Dis. 2016;13:544–50.
Liljebjelke KA, Hofacre CL, Liu T, White DG, Ayers S, Young S, Maurer JJ. Vertical and horizontal transmission of Salmonella within integrated broiler production system. Foodborne Pathog Dis. 2005;2:90–102.
Mead GC, Hudson WR, Hinton MH. Use of a marker organism in poultry processing to identify sites cross-contamination and evaluate possible control measures. British. Poult Sci. 1994;35:345–54.
Min KJ, Hwang IG, Lee SH, Cho JI, Yoon KS. Determination of risk ranking of combination of potentially hazardous foods and foodborne pathogens using a risk ranger. J Fd Hyg Safety. 2011;26:91–9.
National Poultry Improvement Plan, Animal and Plant Health Inspection Service. National poultry improvement plan program standards. In: United States Department of Agriculture. Washington: USDA-APHIS; 2014. p. 30–43.
Popoff MY, Le Minor L. Antigenic formulas of the Salmonella serovars, 7th revision. Pasteur Institute, Paris: World Health Organization Collaborating Centre for Reference and Research on Salmonella; 1997.
Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67.
Russell SM. The effect of airsacculitis on bird weight, uniformity, fecal contamination, processing errors and populations of Campylobacter spp. and Escherichia coli. Poult Sci. 2003;82:1326–31.
Shao D, Shi Z, Wei J, Ma ZA. Brief review of foodborne zoonoses in China. Epidemiol Infect. 2011;139:1497–504.
Smith JL, Fratamico PM. Factors involved in the emergence and persistence of food-borne diseases. J Food Prot. 1995;58:696–708.
Suresh T, Hatha AA, Sreenivasan D, Sangeetha N, Lashmanaperumalsamy P. Prevalence and antimicrobial resistance of Salmonella enteritidis and other salmonellas in the eggs and egg-storing trays from retail markets of Coimbatore, South India. Food Microbiol. 2006;23:294–9.
Van Der Fels-Klerx HJ, Jacobs-Reitsma WF, Van Brakel R, Van Der Voet H, Van Asselt ED. Prevalence of Salmonella in the broiler supply chain in the Netherlands. J Food Prot. 2008;71:1974–80.
vd Giessen AW, Peters R, Berkers PA, Jansen WH, Notermans SH. Salmonella contamination of poultry flocks in The Netherlands. Vet Q. 1991;13:41–6.
Ward LR, Threlfall J, Smith HR, O'Brien SJ. Salmonella enteritidis epidemic. Science. 2000;287:1753–4.
Williams JE, Dillard LH. Salmonella penetration of fertile and infertile chicken eggs at progressive stages of inoculation. Avian Dis. 1968;12:629–35.
Woo YK. Survey on the status of microbial contamination of chicken meats collected from poultry processing plants in nationwide. Korean. J Microbiol. 2007;43:186–92.
Acknowledgements
We thank Youngho Hong, Deokhwan Na, Byoungyoon Kim, and Soowon Choi for their excellent technical assistance.
Funding
This research was supported by a fund (Z-1541781-2013-15-02) by Research of Animal and Plant Quarantine Agency, South Korea.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Author information
Authors and Affiliations
Contributions
JSH analyzed the data and wrote the manuscript. KWS and YBK collected samples and performed serotyping. MSK and CSS performed PFGE analysis. YJL provided the basic format of the study and acted as study team leader. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ha, J.S., Seo, K.W., Kim, Y.B. et al. Prevalence and characterization of Salmonella in two integrated broiler operations in Korea. Ir Vet J 71, 3 (2018). https://doi.org/10.1186/s13620-018-0114-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13620-018-0114-4