Abstract
The expression of T regulatory cells (Foxp3), regulatory (interleukin [IL]-10 and transforming growth factor beta [TGF-β]) and proinflammatory (tumor necrosis factor alpha [TNF-α] and interleukin [IL]-1β) cytokines was quantified using real time polymerase chain reaction (qRT-PCR) in the liver of sheep during early stages of infection with Fasciola hepatica (1, 3, 9, and 18 days post-infection [dpi]). Portal fibrosis was also evaluated by Masson’s trichrome stain as well as the number of Foxp3+ cells by immunohistochemistry. Animals were divided into three groups: (a) group 1 was immunized with recombinant cathepsin L1 from F. hepatica (FhCL1) in Montanide adjuvant and infected; (b) group 2 was uniquely infected with F. hepatica; and (c) group 3 was the control group, unimmunized and uninfected. An overexpression of regulatory cytokines of groups 1 and 2 was found in all time points tested in comparison with group 3, particularly at 18 dpi. A significant increase of the number of Foxp3+ lymphocytes in groups 1 and 2 was found at 9 and 18 dpi relative to group 3. A progressive increase in portal fibrosis was found in groups 1 and 2 in comparison with group 3. In this regard, group 1 showed smaller areas of fibrosis than group 2. There was a significant positive correlation between Foxp3 and IL-10 expression (by immunohistochemistry and qRT-PCR) just as between portal fibrosis and TGF-β gene expression. The expression of proinflammatory cytokines increased gradually during the experience. These findings suggest the induction of a regulatory phenotype by the parasite that would allow its survival at early stages of the disease when it is more vulnerable.
Similar content being viewed by others
Introduction
Fasciolosis, caused by Fasciola hepatica, produces high economic losses to the agricultural sector [1]. This parasite infects a wide range of domestic animals including cattle, sheep, and goats, and the disease has been recognized as an important zoonosis in Africa, Asia, Europa, America, and Oceania [2, 3]. Traditionally, the control of the disease has been based on the use of anthelmintic drugs. Nevertheless, resistance to triclabendazole and other drugs as well as the public concern about the presence of drug metabolites in foodstuff is increasing in numerous countries [4]. Due to this, during the last few years an enormous interest in the development of an immunological method to control the disease has risen [5, 6]. Despite major efforts during the last two decades, the search for an effective vaccine to control this disease has been slow. This is due to the different mechanisms used by F. hepatica to modulate the host immune response making it ineffective to kill the parasites [7,8,9]. One of these mechanisms, like for other helminths, is the expansion of T regulatory cells (Foxp3), facilitating both parasite survival and modulation of tissue damage [10, 11]. Our group has recently shown the expansion of Foxp3+ T cells in the liver and hepatic lymph nodes of sheep and goats infected with F. hepatica [12].
In previous studies, it has been reported that F. hepatica is able to downregulate the Th1 immune response and upregulate the Th2 response at early stages of infection in sheep [13] and mice [14] as well as in chronic stages in cattle [15]. This imbalance towards a Th2 immune profile is mediated through regulatory cytokines and cells that modulate and/or suppress inflammatory responses. The induction of a regulatory environment by the expression of cytokines such as IL-10 and TGF-β has been shown as a common strategy used by parasites and microorganisms to extend their survival [16,17,18]. As a consequence of this regulatory environment, the expression of Foxp3 T cells is increased. Specifically, in F. hepatica infection it has been shown that Foxp3+ lymphocytes play an important role contributing to the parasite survival during the migratory stage [10, 12]. In addition, F. hepatica develops other mechanisms to evade the host’s immune response in early stages in sheep where larvae are able to induce apoptosis of peritoneal leukocytes, allowing the migration of larvae through the peritoneum [19].
In rats, the protective response against F. hepatica has been reported during initial stages of infection [20]. On the other hand, an increase in inducible nitric oxide synthase (iNOS) expression in peritoneal leukocytes has also been reported in goats, suggesting that eosinophils may play an important role in the host response during early stages of infection [21]. For all these reasons, it is of crucial importance to study the host’s immune mechanisms at this stage of infection when the parasite seems to be more vulnerable to the immune response.
The aim of this study was to evaluate the gene expression of regulatory cytokines (IL-10 and TGF-β), proinflammatory cytokines (TNF-α and IL-1β), the transcription factor Foxp3 at different levels (gene and antigenic expression), and portal fibrosis in liver tissue samples from unimmunized and immunized (recombinant cathepsin L1 -FhCL1) sheep during early stages of the infection with F. hepatica.
Materials and methods
Experimental design
The experimental design has been described previously by our group [13]. Briefly, 44 seven-month old female Merino-breed sheep obtained from a liver fluke-free farm were used for this study. All animals were tested monthly for parasite eggs by fecal sedimentation with negative results in all cases. Moreover, prior to the challenge, all animals were tested for serum IgG specific for F. hepatica cathepsin L1 (FhCL1) by enzyme-linked immunosorbent assay ELISA, with negative results in all cases. Animals were housed indoors (100 m2 covered and 100 m2 uncovered facility) and fed with hay and pellets and water ad libitum. The sheep were distributed into three groups: group 1 (n = 20) was immunized subcutaneously with two doses 4 weeks apart with 100 µg rCL1 in 1 mg of Montanide ISA 70 VG (Seppic, Puteaux, France). Recombinant FhCL1 was kindly provided by Prof. John Dalton (Queen’s University Belfast, Belfast, UK). group 2 (n = 20) was unimmunized and infected. group 3 (n = 4) was unimmunized and uninfected.
Groups 1 (immunized and infected) and 2 (unimmunized and infected) were orally infected with 200 metarcercariae of the South Gloucester strain of F. hepatica (Ridgeway Research Ltd, UK) and divided into four subgroups each (n = 5), which were euthanized five animals from group 1 and five animals from group 2 at 1, 3, 9, and 18 days post-infection (dpi). Ten sheep were sacrificed daily (5 from group 1 and 5 from group 2). In all animals, the euthanasia was conducted by intravenous injection of 7 mL of Embutramide 200 mg and Mebezonium iodide 50 mg. No adverse reactions or clinical signs were noted during the experiments. The experiment was approved by the Bioethics Committee of the University of Cordoba (No. 1118) and conducted in accordance with European (2010/63/UE) and Spanish (RD 1201/2005) directives on animal experimentation. The vaccine was non-protective as reported in a previous work [13].
Histopathology
At necropsy, the liver was removed and photographed on the visceral and diaphragmatic aspects for gross evaluation. Liver tissue samples were collected and samples frozen for qRT-PCR examination, while other samples were fixed in 10% neutral buffered formalin for 24 h, then routinely processed and embedded in paraffin wax. Four micron-thick tissue sections were stained with hematoxylin and eosin (H&E) for histopathology. Masson’s trichrome staining (Bio-Optica, Milan, Italy) was used to assess portal fibrosis in liver tissue slides.
Immunohistochemical analysis
An immunohistochemical study was used to assess Foxp3 expression in liver tissue samples. The avidin–biotin–peroxidase method described elsewhere [12] was carried out. Briefly, after hydration of samples, antigen retrieval was carried out by incubating the slides in citric acid (pH 6.0) followed by heating in an autoclave for 10 min at 121 °C. Samples were rinsed twice for 5 min in phosphate buffered saline (PBS) and 5 min in PBS-Tween 80 (Panreac, Barcelona, Spain). Endogenous peroxidase activity was blocked by incubation with 0.3% hydrogen peroxide (Panreac, Barcelona, Spain) in PBS-Tween 80. The slides were then rinsed twice for 5 min in PBS-Tween 80 and incubated with 25% normal goat serum (Vector Laboratories, Burlingame, California, USA) for 1 h at room temperature. An anti-mouse/rat Foxp3 monoclonal antibody (clone FJK-16s, rat IgG2a, eBioscience Inc. San Diego, CA, USA) was diluted 1:100 in PBS containing 10% normal goat serum and applied to the slides overnight as primary antibody. Then, two rinses in 10% normal goat serum were performed prior to a goat anti-rat immunoglobulin serum (Vector Laboratories, Burlingame, California, USA) diluted 1:100 that was applied for 30 min. After three rinses of 7 min in PBS an avidin–biotin–peroxidase complex (Vector Laboratories, Burlingame, California, USA) diluted 1:50 was applied for 1 h. Tissue sections were then washed three times in Tris-buffered-saline (TBS, pH 7.2) and incubated with the vector NovaRED® peroxidase substrate kit (Vector Laboratories, Burlingame, California, USA) for 5 min. Then, samples were rinsed in tap water, lightly counterstained with Mayer’s hematoxylin and mounted with Eukitt® (Freiburg, Germany). Specific primary antibodies were substituted with PBS or non-immune isotype-matched sera as negative control. Hepatic lymph node sections from sheep were used as positive controls.
Morphometry and cell counting
Both portal fibrosis evaluation and Foxp3+ cell counting was done using the biomedical software Image J v.1.51d. In order to measure the fibrotic areas, three liver slides per animal were used and five random microphotographs at 200× magnification were taken for each slide. It was necessary to develop specific macros to calibrate the measure and intensity of positive areas (expressed in µm2) for Masson’s trichrome staining. The number of Foxp3+cells were carried out using three liver slides per animal and five random microphotographs at 400× magnification. In this case, the specific macros developed were carried out to calibrate the appropriate immunostaining intensity and cell size. In both cases, the results were expressed as mean ± SD.
RNA extraction and cDNA synthesis
Samples collected from the left liver lobe were washed in diethylpyrocarbonate (DEPC) biomolecular water and immediately frozen in liquid nitrogen, individually disrupted in liquid nitrogen, and finally stored at −80 °C. Total RNA was isolated from 300 mg of ground tissue homogenized in 1.5 mL of TRIzol® reagent (Ambion Life Technologies, Carlsbad, CA, USA) using a sterilized IKA®T10 basic disperser and then cleaned with the RNeasy® Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s guidelines. Incubation with RNase-free DNase I (Qiagen, Hilden, Germany) for 15 min was included in the protocol. Isolated total RNA was finally incubated at 65 °C for 10 min and kept at −80 °C until used. Concentration and purity of RNA were determined by spectrophotometry. The Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) was used to determine the RNA integrity number (RIN), whose values range from 0 for degraded RNA to 10 for intact RNA [22]. The quality criteria to use RNA samples in the qRT-PCR experiment were: (1) RIN values ≥ 8.5; (2) ratios 260/280 about 2; and (3) absence of gDNA. The iScript™ cDNA Synthesis Kit (BioRad, Hércules, CA, USA) was used to generate cDNAs from 1 μg of total RNA from each sample individually.
Primer design
The primer pairs for gene expression analysis of the targets Foxp3, IL-10, TGF-β, TNF-α, and IL-1β in liver tissue were designed with Oligo 7 software (Colorado Springs, USA) over specific sequences obtained from the GenBank database. In all cases, primer sizes were between 25 and 30 base pair (bp) and lengths of generated amplicons ranged from 99 to 192 bp. To secure primer specificity for target cDNA, the sense and antisense primers were located in different close exons. To obtain high specificity and better performance, primers devoid of hairpin and duplex structure were required to have a high temperature (≥ 68–70 °C) and optimal 3’−∆G (≤ −6 kcal/mol) values for use in two-step 94/68 °C PCR. All primer pairs produced amplicons of the predicted size (Table 1). All PCR products were further verified by nucleotide sequencing.
Absolute quantitation of cytokine transcripts by real-time PCR
The SsoAdvanced™ Universal SYBR® Green Supmermix (BioRad) Kit was used in real-time PCR according to manufacturer’s guidelines. Amplifications were performed in triplicate by using 50 ng of cDNA from each animal and 0.3 µM of each primer in a MyiQ™2 Two Color Real-Time PCR Detection System (BioRad). Cycling conditions consisted of 2 min at 95 °C for Platinum Taq activation followed by 40 two-step cycles for melting (15 s, 95 °C), and annealing/extension (30 s, 70 °C). After 40 cycles, a melting curve analysis was performed (60‒95 °C) to verify the specificity of amplicons. Replicate PCR generated highly reproducible results with standard error of the mean (SEM) < 10% of the mean (< 1% for threshold cycle data). All targets amplified with the same optimal PCR efficiency (100%) and high linearity (r > 0.99) in the range of 20 to 2 × 105 pg of total RNA input. An inter-run calibrator (IRC) RNA sample, with a known number of transcripts of the A170 gene, was introduced in each experiment to guarantee the quality of the retro-transcription and to detect and remove inter-run variation. An absolute calibration curve was generated with an in vitro transcribed RNA containing a known number of copies (Figure 1), as previously described [23, 24]. The number of transcript molecules corresponding to each experimental gene was calculated from the linear regression of the calibration curve.
Absolute standard curve used to calculate the number of copies per pg of total RNA of each mRNA. The absolute standard curve was prepared with an in vitro synthesized RNA corresponding to a 457-nt fragment of the mouse Gapdh transcript (GenBank Database: M32599). Concentration was spectrophotometrically determined by A260 and converted to the number of copies using the molecular weight of the RNA fragment. Serial dilutions (109 to 102 RNA copies) were prepared, retrotranscribed, and amplified by real-time PCR. Primers have been previously described [23]. The standard curve was constructed by plotting the log of starting RNA molecules versus the threshold cycle (Ct). The resulting standard curve is linear (r = 0.998) over 7 orders of magnitude. The efficiency (E) value is calculated from the slope of the standard curve equation, as E = 10[− 1/slope]− 1. The slope of the standard curve indicates that the standard is amplified with 100% efficiency. This standard curve was used to determine the number of copies of each experimental transcript, as exemplified for a Ct = 24.5.
Statistical analysis
The number of mRNA molecules per µg RNA total are shown with averages and SEM. Comparisons of variables between control and infected groups were carried out by using Student’s t test followed by Bonferroni correction for multiple comparisons. For both immunohistochemical and morphometrical studies the results were expressed as mean ± standard deviation (SD). The Kolmogorov–Smirnov test was applied to evaluate if data were normally distributed. Data were analyzed with the non-parametric Kruskall–Wallis multiple comparison test with Dunn’s post hoc test. Correlation studies were estimated using the Spearman’s non-parametric correlation test. For all the statistical tests, significance was stablished with a P value < 0.05. The statistics software used were Sigma Stat 5.1 (San Jose, CA, USA) and GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA, USA).
Results
Liver pathology
Gross hepatic lesions of groups 4 and 6 were typical lesions of chronic fasciolosis: tortuous whitish tracts and hemorrhagic spots, mainly affecting the left hepatic lobe. The severity of gross hepatic lesions from groups 4 and 6 was similar. Gross and histopathological hepatic changes in groups 1 and 2 have been reported previously, with the only significant difference between both groups at 9 dpi when group 1 (immunized and infected) showed lower hepatic lesions than group 2 (unimmunized and infected) [13].
Results of fibrosis measurements in groups 1, 2 and 3 are summarized in Figure 2. A gradual increase of hepatic fibrosis through the experiment was found. This increase was statistically significant (P < 0.0001) in both groups infected (Figure 3) at 9 and 18 dpi compared with group 3 (unimmunized and uninfected). At this timepoint, group 2 (unimmunized and infected) showed higher areas of portal fibrosis than group 1 (immunized and infected) (Figure 2).
Comparison of areas with periportal fibrosis stained with Masson’s trichrome. There was a gradual increase of periportal fibrosis across the different dpis, being statistically significant at 9 and 18 dpi (*) with respect the negative control group (P < 0.0001). The immunized animals showed slightly less fibrosis than those infected at 1 and 3 dpi. However, this decrease was significant at 9 and 18 dpi (+) (P < 0.0001).
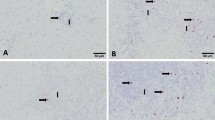
Liver, microphotographs of periportal fibrosis. A Representative picture of periportal fibrosis at 1 and 3 dpi either infected or immunized group. B Periportal fibrosis produced in animals from the infected group at 9 dpi. C Periportal fibrosis of the animals belonging to the immunized group at 18 dpi. D Periportal fibrosis of animals from the infected group at 18 dpi. Masson’s trichrome staining, ×200.
Immunohistochemistry
The anti-Foxp3 mAb yielded a nuclear and/or cytoplasmic immunostaining in lymphocytes mainly located in portal areas (Figure 4) and in a lesser amount in the periphery of granulomas and necrotic foci. The results of the immunohistochemical study of Foxp3 in liver are summarized in Figure 5. There were no significant differences between groups in Foxp3+ cell expression at 1 and 3 dpi. However, a significant increase (P < 0.0001) of Foxp3+ cells was observed at 9 and 18 dpi in groups 1 and 2 compared with group 3. The statistical analysis revealed a strong positive correlation between the number of Foxp3+ T cells in the hepatic inflammatory infiltrates and IL-10 gene expression (r = 0.81; P = 0.0004 and r = 0.88; P < 0.0001 for groups 1 and 2, respectively).
Liver, microphotographs of positive Foxp3 cells. A Positive Foxp3 cells found in both groups, immunized and infected at 1 and 3 dpi. B Positive Foxp3 cells found in the immunized group at 18 dpi. C Positive Foxp3 cells found at 9 dpi and the infected group at dpi. ABC-hematoxylin counterstain, ×200.
Comparison between different groups of positive Foxp3 cells in immunohistochemistry. A significant increase in the expression of Foxp3+ lymphocytes was observed in the infected control group at 9 and 18 dpi (*) with respect to the negative control group and respect to the infected control and immunized groups at 1 and 3 dpi (P < 0.0001). No significant differences were found between the infected control group and the immunized group during the experiment.
Cytokine gene expression
Gene expression levels in liver for Foxp3, IL-10, TGF-β, TNF-α and IL-1β are shown in Figure 6. During the course of infection, Foxp3 expression did not change significantly at 1 and 3 dpi in any of the groups analyzed. Nevertheless, at 18 dpi, groups 1 and 2 showed a significant increase (P < 0.001) in Foxp3 expression compared to group 3. Foxp3 gene expression was higher at 1, 3, and 9 dpi in group 1 compared with group 2. At 18 dpi, group 2 showed higher expression of Foxp3 with respect to the counterparts immunized (Figure 6). There were no statistically significant differences between groups 1 and 2. IL-10 RNA expression increased gradually in both infected groups with respect to the uninfected. This increase was significant from 9 dpi onwards (P < 0.001) in the infected control group and from 3 dpi onwards (P < 0.001) in the infected and immunized group (Figure 6). The immunized and infected group showed higher IL-10 expression than the infected group through all timepoints. Nevertheless, this IL-10 expression increase only was statistically significant at 3 dpi (P = 0.03). There was a significant positive correlation between IL-10 and Foxp3 gene expression for the immunized group (r = 0.68; P = 0.0002) and infected group (P < 0.0001; r = 0.81), respectively.
Gene expression of Foxp3, IL-10, TGF-β, IL-1β, and TNF-α. Each bar represents the mean ± SD of the mRNA molecules/ng of total RNA quantified individually in each of the five animals per experimental condition and sampling time, after three real-time PCR reactions per individual. a for comparison of each treatment with the negative control, and b for comparisons between the infected and infected and immunized groups. The levels of gene expression were increasing gradually throughout the experiment in the transcription factor, foxp3 and two regulatory cytokines studied (IL-10, TGF-β), reaching its higher expression at 18 dpi, and for IL-10 also markedly at 3 and 9 dpi, coinciding with a slight increase in the expression of Foxp3. Foxp3, IL-10, and TGF-β showed significant differences in comparison with the negative control group at 1, 3, and 9 dpi, respectively. At the same time, differences were shown between immunized and infected groups at 1 and 3 dpi in Foxp3 and 3 dpi in IL-10. The expression of proinflammatory cytokines (IL-1β and TNF-α) was gradually increasing along the experience, showing the typical expression of an acute stage of disease.
Gene expression levels for TGF-β showed a significant increase (P < 0.001) in both infected groups at 1, 3, 9, and 18 dpi compared to the uninfected (Figure 6). This increase reached the maximum level at 18 dpi in both infected groups, and the increase was also significantly higher (P < 0.001) than in previous infection stages. There were no differences between the infected group and the immunized and infected group through the experience. The correlation found between TGF-β expression and portal fibrosis also showed a positive correlation for both immunized (P = 0.017; r = 1) and infected (P = 0.017; r = 1) groups.
TNF-α expression exhibited a gradual increase throughout all timepoints. Both infected groups showed a significant increase (P < 0.05) in TNF-α expression in comparison with the uninfected and unimmunized group (group 3), except for the infected/unimmunized group at 1 dpi. Between infected groups, the immunized (group 1) showed a significant increase in the expression of TNF-α at 1, 3, and 9 dpi (P < 0.05) compared with the infected (Figure 6). However, at 18 dpi, the expression was inverse, showing a significant increase (P < 0.001) in the infected group at this timepoint in comparison with the immunized and infected group.
Finally, the expression of IL-1β gradually increased through the experience for both infected groups. This increase was significant at all timepoints compared to the uninfected and immunized group (P < 0.01) and from 9 dpi onward in the infected group (P < 0.001). Notably, the IL-1β expression for both infected groups were two-fold higher at 18 dpi compared with the rest of the timepoints and significantly higher (P < 0.001) than the level in the negative control group. There were no differences between infected groups.
Discussion
The results of the present study revealed a significant increase in T regulatory cells (Foxp3+) detected by immunohistochemistry and gene expression in liver of both infected control (group 2) and immunized (group 1) sheep compared to the uninfected control group at 9 and 18 dpi. These results are in agreement with the significant increase of this cell type described in goats and sheep experimentally infected with F. hepatica at 9 dpi [12] and with the increase of T regulatory cells (Foxp3+) in the abomasum of sheep infected with Teladorsagia circumincta [11] and suggests that both parasites induce expansion of Foxp3+ T cells to modulate the host response facilitating their survival in host tissues. The dynamic of Foxp3 expression detected by immunohistochemistry (IHC) and by PCR was similar in the immunized and infected control groups, which was expected since the vaccine was non-protective. In T. circumcincta infected sheep, expansion of Foxp3+ T cells was associated with increased IL-10 gene expression [11]. The results of the present study revealed a very similar dynamic of IL-10 gene expression and Foxp3 expression of both antigens and gene as well as strong correlation between IL-10 and Foxp3 in the two infected groups, which is in agreement with previous results reported in sheep infected with T. circumcincta [11]. An increase in IL-10 has also been reported in peripheral blood mononuclear cell (PBMC) from sheep experimentally infected with F. hepatica during chronic infection stages, and levels were significantly higher in sheep with high fluke burdens [25]. Cattle naturally infected with F. hepatica also showed increased gene expression for IL-10 in liver tissues compared to uninfected control cattle [26], suggesting a role of this cytokine in modulating the host response facilitating F. hepatica survival during chronic infection. The results of the present study revealed that the role of increases in IL-10 modulating the host response is relevant since the early stages of the hepatic migratory stage.
Hepatic fibrosis is a typical lesion of chronic fasciolosis in sheep [27, 28] and TGF-β inducing SMAD signaling plays an important role in fibrosis progression [29]. The evaluation of portal fibrosis in Masson’s trichrome stained sections in the present study revealed that fibrosis is increased significantly both in the immunized and in the infected control groups at 9 and, particularly, at 18 dpi compared to the uninfected control. The lower fibrosis found at 9 and 18 dpi in group 1 respect to group 2 is in accordance with the higher gross hepatic lesion found at 9 dpi in the group 2 compared to the group 1 [13]. Portal fibrosis is a chronic lesion that take some days to develop, thus since at 9 dpi hepatic lesions from group 2 were more severe than in group 1, this may justify that at 9 and 18 dpi hepatic fibrosis was lower in group 1 than in group 2. The portal fibrosis intensify coincides with the increase of TGF-β gene expression in both infected groups, which occurs from 1 dpi onwards, and it was particularly high at 18 dpi. These results agree with the reported upregulation of the COL1A1 gene induced by TGF-β in PBMCs from F. hepatica infected sheep at 7 dpi [30] and at chronic stages of infection [25, 31]. Researchers have identified increases in the expression of several genes (SMAD3, SMAD4, COL1A1) related to TGF-β and associated with evidence of fibrosis-associated cell types, including fibroblast and stellate cell lines, in acute stages of infection in sheep.
Two of the most participative cytokines in the inflammatory reactions of the innate immune response in mammals are IL-1β and TNF-α, and are present in the acute stages of disease [32, 33]. IL-1β is responsible of the promotion of phagocyte activity favoring the host immune response against infection [34]. On the other hand, TNF-α is an important mediator of proinflammatory mechanisms [35]. A study showed that mice intraperitoneally injected with fatty acid binding protein (Fh12) from F. hepatica and sacrificed at 12 h post-infection, did not express proinflammatory cytokines. And mice injected with Fh12 and subsequently with bacterial lipopolysaccharide (LPS) resulted in significantly reduced levels of TNF-α, likewise in the same experiment they found suppression of the expression of IL-1β and TNF-α in cultivated cells from bone marrow previously treated with LPS and Fh12 [36]. Similar results were found in our study due to the fact that, at 1 dpi, low expression of mRNA for both cytokines were found in comparison with higher levels found at 3, 9, and 18 dpi. It was remarkable that at 1, 3, and 9 dpi gene expression for TNF-α was higher in group 1 (immunized) than in group 2 (infected control), despite the vaccine being non-protective. This finding agrees with the higher TNF-α expression in liver tissues from rats immunized with the myosin regulatory light chain (MRLC) of F. hepatica and partially protected compared to unimmunized rats in chronic infections, coinciding with lower hepatic lesions in immunized rats [37]. By contrast, in the present study, immunized sheep did not show fluke burden reduction, but a reduction of gross hepatic lesions was found at 9 dpi in immunized sheep compared to unimmunized and infected animals [13].
In summary, the results of the present study revealed no consistent differences in proinflammatory and regulatory cytokine expression in liver tissue from immunized (not protected) and unimmunized sheep infected with F. hepatica. The upregulation of Foxp3 jointly with the overexpression of IL-10 and TGF-β in both immunized and non- immunized sheep suggests that F. hepatica induces a modulation of the host response in the very early stages of infection to facilitate the parasite survival at these critical stages of the disease. Finally, the smaller fibrotic areas and lower levels of Foxp3+ cells (IHC and RT-PCR) found in the livers of the immunized and infected group as well the lower gross lesions found particularly at 9 dpi [13] suggest the vaccine induced partial but not sufficient protection. Deeper studies are needed to know the immune mechanisms involved at initial stages of this disease with the aim of developing an effective vaccine.
Abbreviations
- Foxp3:
-
forkhead box P3 transcription factor
- IL10:
-
interleukin 10
- TGFβ:
-
transforming growth factor beta
- TNFα:
-
tumor necrosis factor alpha
- IL1β:
-
interleukin 1
- qRT-PCR:
-
real time polymerase chain reaction
- rCL1:
-
recombinant cathepsin L1
- iNOS:
-
inducible nitric oxide synthase
- FhCL1:
-
Fasciola hepatica cathepsin L1
- ELISA:
-
enzyme-linked immunosorbent assay
- PBS:
-
phosphate buffered saline
- DEPC:
-
diethylpyrocarbonate
- bp:
-
base pair
- RIN:
-
RNA integrity number
- IHC:
-
immunohistochemistry
- PBMC:
-
peripheral blood mononuclear cell
References
Marcos JA, Terashima A, Gotuzo E (2008) Update on hepatobiliary flukes: fascioliasis, opisthorchiasis and clonorchiasis. Curr Opin Infect Dis 21:523–530
Mas-Coma S (2005) Epidemiology of fascioliasis in human endemic areas. J Helminthol 79:207–216
González LC, Esteban JG, Bargues MD, Valero MA, Ortiz P, Náquira C, Más-Coma S (2011) Hyperendemic human fascioliasis in Andean valleys: an altitudinal transect analysis in children of Cajamarca province, Peru. Acta Trop 120:119–129
Fairweather I (2009) Triclabendazole progress report, 2005–2009: an advancement of learning? J Helminthol 83:139–150
Dalton JP, O’Neill S, Stack C, Collins P, Walshe A, Sekiya M, Doyle S, Mulcahy G, Hoyle D, Khazznadji E, Moire N, Brennan G, Mousley A, Kreshchenko N, Maule AG, Donnelly SM (2003) Fasciola hepática cathepsin L-like proteases: biology, function and potential in the development of first generation liver fluke vaccines. Int J Parasitol 33:1173–1181
Piedrafita D, Spithill TW, Smith RE, Raadsma HW (2010) Improving animal and human health through understanding liver fluke immunology. Parasite Immunol 32:572–581
Dalton JP, Robinson MW, Mulcahy G, O’Neill SM, Donnelly S (2013) Immunomodulatory molecules of Fasciola hepatica: candidates for both vaccine and immunotherapeutic development. Vet Parasitol 195:272–285
Toet H, Piedrafita DM, Spithill TW (2014) Liver fluke vaccines in ruminants: strategies, progress and future opportunities. Int J Parasitol 44:915–927
Molina-Hernández V, Mulcahy G, Pérez J, Martínez-Moreno A, Donnelly S, O’Neill SM, Dalton JP, Cwiklinski K (2015) Fasciola hepatica vaccine: we may not be there yet but we’re on the right road. Vet Parasitol 208:101–111
Taylor MD, Van der Werf N, Maizels RM (2012) T cells in helminth infection: the regulators and the regulated. Trends Immunol 33:181–189
McNeilly TN, Rocchi M, Bartley Y, Brown JK, Frew D, Longhi C, McLean L, McIntyre J, Nisbet AJ, Wattegedera S, Huntley JF, Matthews JB (2013) Suppression of ovine lymphocyte activation by Teladorsagia circumcincta larval excretory–secretory products. Vet Res 44:70
Escamilla A, Zafra R, Pérez J, McNeilly TN, Pacheco IL, Buffoni L, Martínez-Moreno FJ, Molina-Hernández V, Martínez-Moreno A (2016) Distribution of Foxp3+ T cells in the liver and hepatic lymph nodes of goats and sheep experimentally infected with Fasciola hepática. Vet Parasitol 230:14–19
Pacheco IL, Abril N, Morales-Prieto N, Bautista MJ, Zafra R, Escamilla A, Ruiz MT, Martínez-Moreno A, Pérez J (2017) Th1/Th2 balance in the liver and hepatic lymph nodes of vaccinated and unvaccinated sheep during acute stages of infection with Fasciola hepatica. Vet Parasitol 238:61–65
Chung JY, Bae YA, Yun DH, Yang HJ, Kong Y (2012) Experimental murine fascioliasis derives early immune suppression with increased levels of TGFβ and IL–4. Korean J Parasitol 50:301–308
Ingale SL, Singh P, Raina OK, Mehra UR, Verma AK, Gupta SC, Mulik SV (2008) Interferon-gamma and interleukin-4 expression during Fasciola gigantica primary infection in crossbred bovine calves as determined by real–time PCR. Vet Parasitol 152:158–161
Hori S, Nomura T, Sakaguchi S (2003) Control of regulatory T cell development by the transcription factor Foxp3. Science 299:1057–1061
Hawrylowicz CM, O’Garra A (2005) Potential role of interleukin-10-secreting regulatory T cells in allergy and asthma. Nat Rev Immunol 5:271–283
Curotto de Lafaille MA, Lafaille JJ (2008) Natural and adaptive FoxP3+ regulatory cells: more of the same or a division of labor? Immunity 29:114–126
Escamilla A, Pérez-Caballero R, Zafra R, Bautista MJ, Pacheco IL, Ruiz MT, Martínez-Cruz MS, Martínez-Moreno A, Molina-Hernández V, Pérez J (2017) Apoptosis of peritoneal leucocytes during early stages of Fasciola hepatica infection in sheep. Vet Parasitol 238:49–53
Van Milligen FJ, Cornelissen JB, Bokhout BA (1999) Protection against Fasciola hepatica in the intestine is highly correlated with eosinophil and immunoglobulin G1 responses against newly excysted juveniles. Parasite Immunol 21:243–251
Zafra R, Pérez-écija RA, Buffoni L, Pacheco IL, Martínez-Moreno A, LaCourse EJ, Perally S, Brophy PM, Pérez J (2013) Early hepatic and peritoneal changes and immune response in goats vaccinated with a recombinant glutathione transferase sigma class and challenged with Fasciola hepatica. Res Vet Sci 94:602–609
Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassman M, Lightfoot S, Menzel W, Granzow M, Ragg T (2006) The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7:3
Prieto-Álamo MJ, Cabrera-Luque JM, Pueyo C (2003) Absolute quantitation of normal and ROS-induced patterns of gene expression: an in vivo real-time PCR study in mice. Gene Expr 11:23–34
Jurado J, Fuentes-Almagro CA, Prieto-Alamo MJ, Pueyo C (2007) Alternative splicing of c-fos pre-mRNA: contribution of the rates of synthesis and degradation to the copy number of each transcript isoform and detection of a truncated c-Fos immunoreactive species. BMC Mol Biol 8:83
Haçariz O, Sayers G, Flynn RJ, Lejeune A, Mulcahy G (2009) IL-10 and TGF-beta1 are associated with variations in fluke burdens following experimental fasciolosis in sheep. Parasite Immunol 31:613–622
Mendes EA, Mendes TA, dos Santos SL, Menezes-Souza D, Bartholomeu DC, Martins IV, Silva LM, Lima Wdos S (2013) Expression of IL-4, IL-10 and IFN-γ in the liver tissue of cattle that are naturally infected with Fasciola hepatica. Vet Parasitol 195:177–182
Meeusen E, Lee CS, Rickard MD, Brandon MR (1995) Cellular responses during liver fluke infection in sheep and its evasion by the parasite. Parasite Immunol 17:37–45
Pérez J, Ortega J, Moreno T, Morrondo P, López-Sández C, Martínez-Moreno A (2002) Pathological and immunohistochemical study of the liver and hepatic lymph nodes of sheep chronically reinfected with Fasciola hepatica, with or without triclabendazole treatment. J Comp Pathol 127:30–36
Cutroneo KR (2007) TGF-β induced fibrosis and SMAD signaling: oligo decoys as natural therapeutics for inhibition of tissue fibrosis and scarring. Wound Rep Reg 15:54–60
Fu Y, Chryssafidis AL, Browne JA, O’Sullivan J, McGettigan PA, Mulcahy G (2016) Transcriptomic study on ovine immune responses to Fasciola hepatica infection. PLoS Negl Trop Dis 10:e0005015
Fu Y, Browne JA, Killick K, Mulcahy G (2017) Network analysis of the systemic response to Fasciola hepatica infection in sheep reveals changes in fibrosis, apoptosis, toll-like receptors ¾, and B cell function. Front Immunol 8:485
Titus RG, Sherry B, Cerami A (1991) The involvement of TNF, IL-1 and IL-6 in the immune response to protozoan parasites. Immunol Today 12:13–16
Eberhard BA, Anderson U, Laxer RM, Rose V, Silverman ED (1995) Evaluation of the cytokine response in Kawasaki disease. Pediatr Infect Dis J 14:199–203
Dinarello CA (1997) Interleukin-1. Cytokine Growth Factor Rev 8:253–265
Goetz FW, Planas JV, MacKenzie S (2004) Tumor necrosis factor. Dev Comp Immunol 28:487–497
Martin I, Cabán-Hernández K, Figueroa-Santiago O, Espino AM (2015) Fasciola hepatica fatty acid binding protein inhibits TLR4 activation and suppresses the inflammatory cytokines induced by LPS in vitro and in vivo. J Immunol 194:3924–3936
Henker LC, Schwertz CI, Lucca NJ, Piva MM, Prior KC, Baska P, Norbury L, Januszkiewicz K, Dezen D, Duarte MMMF, Moresco RN, Bertagnolli da Rosa L, Mendes RE (2017) Immune protection conferred by recombinant MRLC (myosin regulatory light chain) antigen in TiterMax Gold® adjuvant against experimental fasciolosis in rats. Vaccine 35:663–671
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiment: AMM and JP. Performed the experiment: ILP, MJB, MTR, RZ, RPC, VMH. Analyzed the data of qRT-PCR: NA. Laboratory work: ILP, NMP. Analyzed statistics data: NA, RZ. Wrote and revised the paper: ILP, JP, NA, RZ, AMM. All author read and approved the final manuscript.
Acknowledgements
This work was supported by EU Grant (H2020-635408-PARAGONE) and National grant (AGL2015-67023-C2-1-R). We thank Prof. John P. Dalton, Queen’s University Belfast, Northern Ireland, for providing recombinant FhCL1. Sequencing analysis and quality analysis of RNA were carried out by the Central Services for Research of the University of Córdoba (SCAI).
Ethics approval and consent to participate
The experiment was approved by the Bioethics Committee of the University of Cordoba (No. 1118) and conducted in accordance with European (2010/63/UE) and Spanish (RD 1201/2005) directives on animal experimentation.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Pacheco, I.L., Abril, N., Zafra, R. et al. Fasciola hepatica induces Foxp3 T cell, proinflammatory and regulatory cytokine overexpression in liver from infected sheep during early stages of infection. Vet Res 49, 56 (2018). https://doi.org/10.1186/s13567-018-0550-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-018-0550-x