Abstract
Campylobacter jejuni is the leading cause of bacterial food-borne gastroenteritis worldwide and human infections are frequently associated with handling and consumption of contaminated poultry. The polysaccharide capsule of C. jejuni plays important roles in colonisation of the chicken gut, invasion of epithelial cells and serum resistance and is subject to modification with O-methyl phosphoramidate (MeOPN) in most strains. In this study, the cytokine responses of mouse bone marrow-derived macrophages (mBMMs), chicken bone marrow-derived macrophages (chBMMs) and human monocyte-derived macrophages (hMDMs) were measured following infection with C. jejuni 11168H wild-type (WT) or isogenic mutants lacking either the capsule (Δcj1439) or its MeOPN modification (Δcj1417). Consistent with previous observations using murine bone marrow-derived dendritic cells, mutants lacking the capsule or MeOPN elicited enhanced transcription of IL-6 and IL-10 in mBMMs compared to wild-type C. jejuni. However, the lack of capsule and MeOPN did not alter IL-6 and IL-10 expression in chBMMs and hMDMs compared to C. jejuni WT. Phagocytosis assays showed the acapsular mutant was not impaired in uptake or net intracellular survival after phagocytosis in both chicken and human macrophages; however, the phagocytosis of the MeOPN mutant was significantly decreased in both chicken and human macrophages. In conclusion, differences in the response of macrophages of varying host origin to Campylobacter were detected. The absence of MeOPN modification on the capsule of C. jejuni did not alter the levels of innate cytokine expression in both chicken and human macrophages compared to the 11168H WT, but affected phagocytosis by host macrophages.
Similar content being viewed by others
Introduction
Campylobacter jejuni is the leading cause of bacterial food-borne gastroenteritis worldwide [1]. Approximately 70 353 laboratory-confirmed cases of human infection were recorded by national surveillance in the United Kingdom from February 2014 to February 2015 (Zoonoses summary report UK 2014), however the true burden of infection in the community is expected to be far higher [2]. Symptoms of infection in humans vary from mild watery diarrhoea to severe bloody diarrhoea. Disease is usually self-limiting and symptoms last for 5–7 days, but in some cases the infection may be complicated by severe sequelae [3, 4]. Complications of infection include septicaemia and inflammatory neuropathies such as Guillain–Barré syndrome that affect approximately 1/1000 cases [5, 6].
The human immune response to C. jejuni, including the role of pattern recognition receptors and bacterial agonists of innate and adaptive immunity, remains poorly understood. As in other enteric infections, innate immune cells are likely to be involved in the initial response to C. jejuni infection [7, 8]. Upon C. jejuni infection, human monocytic cell line THP-1 and monocyte-derived dendritic cells (DCs) secrete a broad range of cytokines including IL-1β, IL-8, IL-10, IFN-γ, and TNF-α [9, 10]. Additionally, live C. jejuni induces a higher cytokine response in DCs than killed bacteria [11]. The pathogen recognition receptors, toll-like receptors (TLR) 2 and 4 have been shown to be involved in signalling and cytokine expression upon infection with C. jejuni in vitro using knock-out or knock-down technology [12, 13]. A luciferase-based TLR reporter assay, however, indicated that live C. jejuni did not directly activate human TLR2, TLR4, and TLR5 [14]. Human monocytes/macrophages also efficiently kill C. jejuni [15].
Chickens are the key reservoir of human Campylobacter infection and up to 50–70% of human cases have been attributed to the handling and consumption of contaminated broiler meat [3, 4]. Campylobacter colonization of chickens is widespread and results in long-term persistence and shedding [16, 17]. Although large numbers of bacteria are in close contact with the epithelial cell layer, primarily in the large blind caeca (where C. jejuni number may exceed 109 CFU/g), no haemorrhagic lesions, caecal dilation and clinical signs of intestines are seen in most colonised birds [18] in clear contrast to humans who develop severe clinical symptoms of diarrhoea. In terms of immune responses, several reports have shown induction of immune-associated gene expression after Campylobacter colonization of chickens. Indeed, evidence exists that C. jejuni can elicit gut pathology, pro-inflammatory responses and impair weight gain in some, but not all, commercial broiler lines [19]. Experimental infection of a commercial broiler line A1 elicits expression of the pro-inflammatory chemokines CXCLi1, CXCLi2 and IL-1β in the ceca at 2 and 5 days post-infection. Moreover, the infected A1 birds developed diarrhoea at 12 days post-infection [19]. Another study reported that experimental infection of a commercial broiler line (Ross 308) with C. jejuni leads to increased mRNA levels of TLR4 and TLR21 in peripheral blood leukocytes, but decreased transcription of other antimicrobial peptide genes in response to C. jejuni infection [20]. Additionally, C. jejuni infection increases the expression of pro-inflammatory cytokines (IL-1β and IL-6) and chemokines (CXCLi1 and CXCLi2) both in vitro as well as in vivo in ileum and caecum [21,22,23]. Infection of C. jejuni leads to an increased number of circulating monocytes/macrophages in commercial broiler chickens (Ross 308) [20], and an increased number of heterophils are recruited in the ileum and caeca of specific pathogen-free chickens challenged with C. jejuni [23]. Similar to human, chicken peritoneal macrophages are able to ingest and kill C. jejuni effectively in the in vitro assay [24].
The analysis of the genome sequence of C. jejuni NCTC 11168 revealed that approximately 8% of its predicted coding sequences are devoted to the biosynthesis of sugar moieties and bacterial surface glycans including lipo-oligosaccharide (LOS), capsular polysaccharide (CPS) and N- and O-linked glycosylation systems [25]. The CPS plays important roles for both serum resistance and invasion of epithelial cells [26]. The CPS produced by C. jejuni strains is structurally complex and highly variable, due in part to the addition of phase-variable modifications such as O-methyl, ethanolamine, aminoglycerol and O-methyl phosphoramidate (MeOPN) groups [27,28,29]. Earlier studies reported that approximately 70% of C. jejuni isolates have MeOPN modification, which has important roles in invasion of epithelial cells and serum resistance [26, 30]. The loss of MeOPN modification leads to increased invasion of epithelial cells compared to the wild-type (WT) strain but with no effect on adherence [30]. A capsule-deficient mutant (Δcj1439) showed significantly decreased adherence and invasion to epithelial cells compared to C. jejuni WT [30, 31]. While the capsule plays a key role in colonisation of the chicken gut [32], the absence of MeOPN modification reportedly does not affect colonisation of C. jejuni in the chicken [20], albeit modest attenuation of a MeOPN mutant was observed in piglets [30]. In mouse bone marrow-derived dendritic cells (mBMDCs), both capsule (Δcj1439) and MeOPN (Δcj1417) mutants significantly enhances IL-6 and IL-10 mRNA and protein expression compared to WT of C. jejuni [33].
In this study, the effect of the capsule and its MeOPN modification on the cytokine responses of macrophages from mice, chickens and human to C. jejuni was examined. Both capsule (Δcj1439) and MeOPN (Δcj1417) mutant strains did not alter cytokine responses in chicken and human macrophages, whereas enhanced cytokine responses were observed in mBMMs by the capsule and MeOPN deficient mutants. Furthermore, the lack of MeOPN modification reduced bacterial uptake, but not net intracellular survival, in chicken and human macrophages compared to C. jejuni WT.
Materials and methods
Animals
Commercial Novogen brown layers and C57/B16 mice were housed in premises licensed under a UK Home Office Establishment License in full compliance with the Animals (Scientific Procedures) Act 1986 and the Code of Practice for Housing and Care of Animals Bred, Supplied or Used for Scientific Purposes. Requests for animals were approved by the local Animal Welfare and Ethical Review Board and animals were humanely culled in accordance with Schedule 1 of the Animals (Scientific Procedures) Act 1986.
Isolation and culture of mouse bone marrow-derived macrophages (mBMMs)
Vials of frozen C57BL/6 mouse bone marrow were kindly provided by Dr Hu (The Roslin Institute), following recovery by standard methods. The mBMMs were differentiated following the protocol of Irvine et al. [34] in the presence of rhCSF1. Briefly, the bone marrows were cultured on 90-mm plastic petri dishes with RPMI 1640 (Sigma) supplemented with 10% (v/v) FBS (Gibco), 2 mM l-glutamine, 100 U/mL Penicillin, 100 µg/mL Streptomycin (Gibco) and recombinant human colony-stimulating factor-1 (rhCSF1, a gift from Chiron, Emeryville, CA, USA, 1 × 104 U/mL as final concentration) for 7 days for differentiation into macrophages [34]. At day 4 post-seeding, 10 mL of fresh culture media with 104 U/mL rhCSF1 was added per petri dish. At 7 days post-seeding, the differentiated mBMMs were harvested using 10 mM EDTA, and re-plated at a concentration of 1 × 106 cells/well in 6-well plates for inoculation with bacterial strains or agonists and analysis of gene expression. The cells were cultured for 24 h in a humidified 5% CO2 atmosphere prior to stimulation.
Isolation and culture of chicken bone marrow-derived macrophages (chBMMs)
Chicken macrophages were differentiated from bone marrow isolated from embryos at day 20 of development. Bone marrow was harvested by flushing femurs and tibias, and washed once with chilled RPMI 1640 medium. Approximately 1.0 × 107 bone marrow cells were cultured on 90-mm plastic petri dishes as previously described [35] in RPMI 1640 medium supplemented with 10% (v/v) FBS, 10 U/mL Penicillin, 10 μg/mL Streptomycin, 2 mM l-glutamine and 350 ng/mL recombinant chicken CSF1 (rchCSF1) at 41 °C in a humidified 5% CO2 atmosphere for 7 days to allow macrophage differentiation. The media was changed on 2 and 4 days post-seeding. At 7 days post-seeding, the differentiated macrophages were harvested using 30 mM EDTA, and re-plated at a concentration of 2 × 106 cells/well in 6-well plates and 5 × 105 cells/well in 24-well plates for gene expression studies and phagocytosis assays, respectively. The cells were cultured for 24 h prior to stimulation.
Isolation and culture of human CD14+ monocyte-derived macrophages (huMDMs)
Human CD14+ mononuclear cells were isolated from blood of consenting donors under ethical approval from Lothian Research Ethics Committee (11/AL/0168). Briefly, the buffy coat was isolated from whole blood by centrifugation, followed by isolation of mononuclear cells by gradient separation using Lymphoprep (Stemcell Technologies). The isolated mononuclear cells were washed, and CD14+ monocytes were positively selected and purified using MACS with CD14 microbeads (10 µL per 107 cells; Miltenyl Biotec). The purified CD14+ monocytes were directly seeded and differentiated on either 6-well plates with 1 × 106 cells/well for gene expression studies or 24-well plates with 5 × 105 cells/well for phagocytosis study, and cultured with RPMI 1640 (Sigma) supplemented with 10% (v/v) FBS (GE Healthcare), 2 mM l-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin (Sigma), and rhCSF1 (1 × 104 U/mL as final concentration) [34] for 7 days for differentiation into macrophages and achieved a comparable density at the time of inoculation to the avian and murine macrophage cultures. Infections were performed on day 8.
Bacterial strains, culture and determination of bacteria concentration
Bacterial strains used in this study are C. jejuni 11168H (WT), an isogenic Δcj1417 mutant lacking phosphoramidate side branch (MeOPN) [36], and acapsular Δcj1439 mutant [37] (kindly provided by Prof. Brendan Wren, London School of Hygiene & Tropical Medicine). All bacteria were cultured under microaerophilic conditions (5% O2, 5% CO2, and 90% N2) at 41 °C as described previously [38]. Initially, cultures were grown on modified charcoal-cephoperazone-deoxycholate agar (mCCDA; Oxoid) plates for 48 h, supplemented with selective supplement containing with cefoperazone (32 mg/L) and amphotericin B (10 mg/L; Oxoid), in the presence of kanamycin (50 µg/mL as final concentration) for mutant strains. A single colony from the initial culture was selected and inoculated to 10 mL of Mueller–Hinton broth (Oxoid), followed by additional culture for 24 h at 41 °C with 400 rpm shaking. Twenty-four hours post-inoculation, morphology and motility of the bacteria was observed using KOVA Glasstic® slide with grid (Kova), with only spiral-shaped and rapidly motile cultures being used. Absorbance was measured at 600 nm wavelength. For live bacteria, the cultured bacteria were collected by centrifugation at 13 000 × g for 10 min. The bacterial pellets were washed with sterile PBS once, and then resuspended with PBS. The number of live bacteria used for stimulation was subsequently verified by serial dilution and plating to establish viable bacterial cell count.
To fix the bacteria, the cultured bacteria were pelleted by centrifugation at 13 000 × g for 10 min. The bacteria were resuspended in 200 μL of sterile PBS, and mixed with an equal volume of 4% (w/v) paraformaldehyde in PBS for 1 h at room temperature. The bacteria were washed 3 times with PBS prior to incubation with macrophages. Bacterial fixation was confirmed by microscopic observation and plating to establish elimination of viability.
Stimulation of macrophages with C. jejuni
The chBMMs and mBMMs were harvested and re-plated in 6-well plates 24 h prior to stimulation. The live and fixed bacteria were diluted with RPMI 1640 medium supplemented with 10% (v/v) FBS, 2 mM l-glutamine and either rchCSF1 or rhCSF1, and were added to the prepared cells at MOIs of 50 or 100 for 3 or 6 h for subsequent analysis. For huMDMs, diluted bacteria were added directly into the huMDMs differentiated for 8 days. Lipopolysaccharide from Salmonella minnesota R595 (100 ng/mL as final concentration; InvivoGen) [39] was used as a positive control and PBS in the culture media as negative control.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from the stimulated macrophages using the RNeasy Mini Spin Column (Qiagen) and 1 μg of total RNA was reverse transcribed with cDNA synthesis kit (Bio-Rad), followed by 1:5 dilution for target transcripts and 1:100 dilution for 28S ribosomal RNA. To measure mRNA level from the chBMMs, 2 μL of diluted cDNA was used with TaqMan® Fast Universal PCR Master Mix (Applied Biosystems, UK) and TaqMan primer and probes (Table 1). For the mRNA levels of human or mouse macrophages, 2 μL of diluted cDNA was mixed with Fast SYBR® Green Master Mix (Applied Biosystems) and specific primer sets (Table 1). All qRT-PCR was performed by Applied Biosystems 7500 Fast real-time PCR system. Cycle thresholds were normalised to internal control (28 s for chicken and human and β-actin for mouse) and calibrated to a medium-treated control sample for relative quantification using the ΔΔCt method [40]. The results were presented as a fold change.
Phagocytosis assay
The assay was performed as described previously [41], with slight modifications. Twenty-four hours prior to stimulation, 5 × 105 cells/well of chBMMs were seeded into 24-well plates. For huMDMs, the purified CD14+ monocytes were seeded on 24-well plates at a density of 5 × 105 cells/well. The prepared cells were inoculated at an MOI of 50 for 30 min without antibiotics. Then, the cells were washed three times with PBS, followed by treatment with gentamicin (200 µg/mL as a final concentration) for 30 min to kill extracellular bacteria. The cells were washed three times with PBS, incubated for 0 or 3 h with RPMI 1640 supplemented with 10% (v/v) FBS, 20 µg/mL of gentamicin and either rchCSF1 or rhCSF1, and subsequently lysed with 0.5% Triton X-100 on ice. The collected lysates were prepared by tenfold serial dilution with PBS and plated on mCCDA plates to determine intracellular bacteria. The colonies were counted, followed by the calculation of the CFU/mL and the percentage of killed bacteria to measure net intracellular survival.
Statistical analysis
All data were analysed by Student’s t test or one-way analysis of variance (ANOVA) using program R [42], and significant differences between groups were considered significant by Tukey’s honest significant difference (HSD) test at P < 0.05 (confidence level = 95%).
Results
Capsule and MeOPN mutants of C. jejuni elicit enhanced cytokine responses in mBMMs
Rose et al. [33] previously described the induction of differential responses in murine dendritic cells using the same strains of C. jejuni. To ensure that the bacteria used in our study would give similar responses, we repeated the study of Rose and stimulated mBMDCs at a multiplicity of infection (MOI) of 50 with live or fixed wild-type (WT) C. jejuni 11168H or isogenic capsule (Δcj1439) and MeOPN (Δcj1417) mutants for 3 h. We confirmed that mBMDCs expressed significantly enhanced IL-6 and IL-10 mRNA levels after stimulation with the capsule and MeOPN deficient mutants of C. jejuni (see Additional file 1).
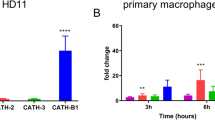
After confirming that the bacteria induced similar responses to previously published data [33], we then stimulated mBMMs with the same strains of C. jejuni under the same condition to examine the effect of capsular and its modification on macrophage responses. mBMMs expressed significantly higher mRNA levels of IL-6 and IL-10 after stimulation with the capsule and MeOPN deficient mutants of C. jejuni compared to the C. jejuni WT (Figure 1). The expression levels of the cytokines did not differ after stimulation with live or fixed C. jejuni.
Campylobacter jejuni capsule and MeOPN mutants elicit an enhanced cytokine responses in mBMMs. Live or fixed C. jejuni 11168H (WT), Δcj1417 (MeOPN) and Δcj1439 (capsule) mutants were incubated with mBMMs at MOI 50 for 3 h. Salmonella LPS was used as a positive control, while PBS in the medium was used as a negative control. Graphs show representative results of two independent experiments and error bars are the standard deviation of two independent experiments. Asterisks indicate a statistically significant difference of mRNA levels of inflammatory-related cytokines compared to C. jejuni WT (*P < 0.05; **P < 0.01).
Cytokine responses of chBMMs and huMDMs to C. jejuni are not affected by the capsule of MeOPN modification
To examine the role of the capsule and its MeOPN modification on the innate immune response of chicken macrophages to C. jejuni, live or fixed C. jejuni WT and capsule (Δcj1439) and MeOPN (Δcj1417) mutants were incubated with chBMMs for 3 and 6 h at an MOI of 50, and transcripts of inflammation-related signature cytokines were measured (Figure 2). The C. jejuni WT, capsule (Δcj1439) and MeOPN (Δcj1417) mutants all significantly induced transcription of the pro-inflammatory cytokines IL-1β and IL-6, the chemokine CXCLi1, and anti-inflammatory cytokine IL-10, compared to the negative control incubated with PBS (Figure 2). A similar induction of responses was observed with live or fixed bacterial cells (Figure 2). However, in contrast to observations with murine cells, no statistically significant differences in the mRNA levels of IL-1β, IL-6, CXCLi1 and IL-10 were detected after stimulation with the WT or mutant strains of C. jejuni. Increasing the MOI to 100 did not result in significantly different transcript levels for the cytokines, either across the strains or time intervals measured (see Additional file 2).
Wild-type and capsule or MeOPN deficient strains of C. jejuni induce similar cytokine responses in chBMMs. Live or fixed C. jejuni 11168H (WT), Δcj1417 (MeOPN) and Δcj1439 (capsule) mutants were incubated with chBMMs at MOI 50 for 3 and 6 h. Salmonella LPS (100 ng/mL) was used as a positive control and mRNA levels of target genes were presented as folder changes compared to PBS in the medium (a negative control). Graphs show representative results of three independent experiments and error bars are standard deviation of three independent experiments. Asterisks indicate a statistically significant difference of mRNA levels of inflammatory-related cytokines compared to PBS in the medium (*P < 0.01; **P < 0.001).
Since TLR4 can trigger inflammatory cytokine response in mammalian innate immune cells by binding to LOS of C. jejuni [10, 43], changes in chicken TLR4 transcript levels upon the stimulation with WT and mutant C. jejuni strains were investigated (see Additional file 3). Stimulation of chBMMs with Salmonella LPS for 3 h led to the downregulation of TLR4 mRNA compared to the cells incubated with medium, as previously reported [44, 45]. Expression of TLR4 mRNA was significantly decreased following stimulation with all three strains of C. jejuni; however there was no difference of chicken TLR4 transcript levels observed among WT, capsule (Δcj1439) and MeOPN (Δcj1417) mutants of C. jejuni regardless of whether live or fixed bacterial cells were used (see Additional file 3).
Since C. jejuni is a zoonotic bacterium with relatively contrasting symptoms in chicken and human, human CD14+ MDMs were cultured and stimulated with an MOI 50 of live or fixed WT or mutant C. jejuni for 3 h to compare inflammatory responses with macrophages or avian and murine origin (Figure 3). All three strains of C. jejuni elicited expression of IL-6 mRNA in huMDMs regardless of the bacteria being live or fixed, but no significant differences between WT and capsule (Δcj1439) or MeOPN (Δcj1417) mutants were found. Unlike IL-6, IL-10 transcription in huMDM were not affected by neither Salmonella LPS nor the different strains of C. jejuni.
Wild-type and capsule or MeOPN deficient strains C. jejuni induce similar cytokine responses in huMDMs. Live or fixed C. jejuni 11168H (WT), Δcj1417 (MeOPN) and Δcj1439 (capsule) mutants were incubated with huMDMs at MOI 50 for 3 h. Salmonella LPS was used as a positive control, and mRNA levels of target gene was presented as folder changes compared to PBS in the medium (a negative control). Graphs show representative results of three individual donors (two individuals for fixed C. jejuni). Error bars are standard deviation of three (two) independent experiments. Asterisks indicate a statistically significant difference of mRNA levels for the indicated inflammation-related cytokines compared to PBS in the medium (*P < 0.01).
Reduced number of intracellular C. jejuni in chicken and human macrophages due to the lack of MeOPN modification
To investigate the effect of the capsule and MeOPN modification on C. jejuni uptake and net intracellular survival in both chicken and human macrophages, a phagocytosis assay was performed (Figure 4). Bacterial recoveries, after gentamicin treatment to kill extracellular bacteria, were used to quantify initial phagocytosis (presented as 0 h) and net intracellular survival (3 h after removal of gentamicin). In both chBMMs and huMDMs, there was no statistically significant differences in the number of C. jejuni taken up (T0) between the wild-type strain and acapsular mutant in macrophages. However, the lack of MeOPN modification led to a significantly reduced uptake (P < 0.03) of intracellular C. jejuni in chBMMs and huMDMs compared to the WT and acapsular mutant (Figure 4A). The percentage of bacteria surviving at 3 h post-incubation revealed no significant difference of net intracellular survival between WT and MeOPN mutants in chBMMs and huMDMs (Figure 4B). The acapsular mutant showed a slight but significant increase in the percentage of bacteria killed in huMDMs, but not chBMMs, 3 h post-incubation compared to WT of C. jejuni.
The lack of MeOPN modification affects the number of intracellular C. jejuni in macrophages. Chicken or human macrophages were incubated with C. jejuni 11168H (WT), Δcj1417 (MeOPN) and Δcj1439 (capsule) mutant at MOI 50 for 30 min. Intracellular bacteria were assessed by a gentamicin protection assay, and counted at 0 h after removal of gentamicin (T0, open bar) and after a further 3 h (T3, closed bar) of incubation. The average number of colonies at T0 and T3 are presented as Log10 CFU/mL (A). The percentage of killed bacteria was calculated using the average number of colonies at T0 and T3 (B). Each bar represents mean of three independent experiments and error bars are the standard deviation of three independent experiments. Asterisks indicate a statistically significant difference of the number of survived bacteria (A) and the percentage of killed bacteria (B) in different treatment (P < 0.05).
Discussion
There has been concerted effort to better understand the differences between the avian and human immune response to C. jejuni infection [9, 10, 14, 21]. Recently, important roles for the capsule and its MeOPN modification have been demonstrated in cell adherence, invasion, colonisation, serum resistance and host immune responses [26, 30, 31, 33]. In this study, we compared the cytokine responses of macrophages of different host origin known to differ in their symptoms upon infection with C. jejuni. The phagocytic and killing capacity of chicken and human macrophages to C. jejuni and capsule or MeOPN deficient mutants were analysed to better understand these differential responses observed in vivo. Although macrophages are not the first cells to contact C. jejuni during the infection in mucosal environment, intestinal and colonic macrophages are part of the first line defence mechanism since they are located strategically underneath the epithelial lining in close proximity to large numbers of luminal bacteria and antigenic stimuli. These gut macrophages regulate inflammatory responses to bacteria and antigens that cross the epithelium, protect the mucosa against harmful pathogen and scavenge dead cells and foreign debris by crosstalking with adjacent intestinal epithelial cells through TLR-dependent signalling pathway [46, 47]. Additionally, a recent study reported crosstalk between intestinal epithelial cells and macrophages mediated through TLR4, resulting in the production of IL-10 from human intestinal epithelial cells [48].
Using the current WT, capsule (Δcj1439) and MeOPN (Δcj1417) strains of C. jejuni, we were able to reproduce the enhanced expression of IL-6 and IL-10 mRNA levels in mBMDCs by capsule and MeOPN deficient strains of C. jejuni compared to the WT strain, as reported by Rose et al. [33]. We also found that mBMMs incubated with the mutant strains expressed elevated levels of IL-6 and IL-10 mRNA compared to the C. jejuni WT, suggesting a potential role for capsule and MeOPN modification in bacterial evasion of mouse innate immune responses. In the context of zoonosis however, it is more relevant to consider the response of avian and human cells to C. jejuni infection and the role of bacterial factors in this process. For this purpose, the mRNA levels of IL-1β, IL-6 and CXCLi1 (pro-inflammatory cytokine/chemokine) and IL-10 (anti-inflammatory cytokine) were measured to examine the immune response of chicken, based on Smith et al. [21]. As markers of human responses, the expression of IL-6 and IL-10 mRNA was measured based on Rose et al. [33]. All three strains of C. jejuni significantly induced mRNA levels of inflammation-related signature genes in both chicken and human macrophages, indicating that C. jejuni is highly immunogenic to both avian and human primary macrophages. These results are very similar with early reports that C. jejuni is highly immunogenic to both human and mouse macrophage cell lines [14, 20]. However, neither capsule nor MeOPN deficient mutants altered the cytokine responses in chicken and human macrophages, when compared to WT, in contrast to the responses of mouse macrophages, indicating that neither of structures is vital for the induction of an immune response in human and avian cells.
All three strains of C. jejuni induced significant mRNA levels of IL-10 in chBMMs and mBMMs compared to PBS in the medium (as negative control). However, there was no significant increase in IL-10 transcripts in huMDMs observed by C. jejuni WT, capsule (Δcj1439) and MeOPN (Δcj1417) mutants. In fact, the expression levels of IL-10 mRNA in huMDMs from all three donors was relatively high compared to chBMMs and mBMMs, although the high expression of IL-10 mRNA did not appear to affect the expression of IL-6 during the stimulation with C. jejuni. Except for the different origin of macrophages between bone marrow and monocytes, there is no obvious explanation for the different responses of IL-10 mRNA levels in huMDMs to C. jejuni compared to chBMMs and mBMMs.
It has been reported that the capsule (Δcj1439) mutant showed significantly decreased adherence and invasion to the epithelial cells [30, 31], while the MeOPN (Δcj1417) mutant has shown increased invasion (but no defect in adherence) to epithelial cells compared to wild-type strains [30]. Additionally, the lack of MeOPN modification, as well as capsule, leads to the significantly decreased serum resistance, indicating a potentially protective role for MeOPN modification against the innate immune system [30]. Our phagocytosis study showed decreased C. jejuni uptake in the absence of MeOPN modification in chicken and human macrophages compared to C. jejuni WT; however, the lack of MeOPN modification did not affect net intracellular survival in chicken and human macrophages. On the other hand, the capsule did not affect bacterial uptake or net intracellular survival at least at the time intervals sampled.
In summary, chicken and human macrophages showed similar responses to C. jejuni WT, capsule (Δcj1439) and MeOPN (Δcj1417), unlike mouse macrophages. The absence of MeOPN modification on the capsule of C. jejuni did not alter the cytokine responses, as well as net intracellular survival in chicken and human macrophages compared to C. jejuni 11168H, but it affected C. jejuni uptake by chicken and human macrophages. The study indicates that extrapolation of findings regarding the potential contribution of candidate virulence factors and host responses to pathogenesis may not always be reproduced across comparable cell types originating from other hosts.
Abbreviations
- DCs:
-
dendritic cells
- TLR:
-
toll-like receptors
- LOS:
-
lipo-oligosaccharide
- CPS:
-
capsular polysaccharide
- MeOPN:
-
O-methyl phosphoramidate
- mBMDCs:
-
mouse bone marrow-derived dendritic cells
- mBMMs:
-
mouse bone marrow-derived macrophages
- chBMMs:
-
chicken bone marrow-derived macrophages
- huMDMs:
-
human monocytes-derived macrophages
- rhCSF1:
-
recombinant human colony-stimulating factor-1
- rchCSF1:
-
recombinant chicken colony-stimulating factor-1
- mCCDA:
-
modified charcoal-cephoperazone-deoxycholate agar
- MOI:
-
multiplicity of infection
References
Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, Praet N, Bellinger DC, De Silva NR, Gargouri N, Speybroeck N, Cawthorne A, Mathers C, Stein C, Angulo FJ, Devleesschauwer B, World Health Organization Foodborne Disease Burden Epidemiology Reference Group (2015) World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med 12:e1001923
Tam CC, Rodrigues LC, Viviani L, Dodds JP, Evans MR, Hunter PR, Gray JJ, Letley LH, Rait G, Tompkins DS, O’Brien SJ, Committee IIDSE (2012) Longitudinal study of infectious intestinal disease in the UK (IID2 study): incidence in the community and presenting to general practice. Gut 61:69–77
Dasti JI, Tareen AM, Lugert R, Zautner AE, Gross U (2010) Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int J Med Microbiol 300:205–211
Epps SV, Harvey RB, Hume ME, Phillips TD, Anderson RC, Nisbet DJ (2013) Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. Int J Environ Res Public Health 10:6292–6304
Tam CC, O’Brien SJ, Petersen I, Islam A, Hayward A, Rodrigues LC (2007) Guillain–Barre syndrome and preceding infection with campylobacter, influenza and Epstein–Barr virus in the general practice research database. PLoS One 2:e344
Nyati KK, Prasad KN, Rizwan A, Verma A, Paliwal VK (2011) TH1 and TH2 response to Campylobacter jejuni antigen in Guillain–Barre syndrome. Arch Neurol 68:445–452
Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P (2001) Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol 2:361–367
Bereswill S, Fischer A, Plickert R, Haag LM, Otto B, Kuhl AA, Dasti JI, Zautner AE, Munoz M, Loddenkemper C, Gross U, Gobel UB, Heimesaat MM (2011) Novel murine infection models provide deep insights into the “ménage à trois” of Campylobacter jejuni, microbiota and host innate immunity. PLoS One 6:e20953
Jones MA, Totemeyer S, Maskell DJ, Bryant CE, Barrow PA (2003) Induction of proinflammatory responses in the human monocytic cell line THP-1 by Campylobacter jejuni. Infect Immun 71:2626–2633
Hu L, Bray MD, Osorio M, Kopecko DJ (2006) Campylobacter jejuni induces maturation and cytokine production in human dendritic cells. Infect Immun 74:2697–2705
Rathinam VA, Hoag KA, Mansfield LS (2008) Dendritic cells from C57BL/6 mice undergo activation and induce Th1-effector cell responses against Campylobacter jejuni. Microbes Infect 10:1316–1324
Friis LM, Keelan M, Taylor DE (2009) Campylobacter jejuni drives MyD88-independent interleukin-6 secretion via Toll-like receptor 2. Infect Immun 77:1553–1560
Rathinam VA, Appledorn DM, Hoag KA, Amalfitano A, Mansfield LS (2009) Campylobacter jejuni-induced activation of dendritic cells involves cooperative signaling through Toll-like receptor 4 (TLR4)-MyD88 and TLR4-TRIF axes. Infect Immun 77:2499–2507
de Zoete MR, Keestra AM, Roszczenko P, van Putten JP (2010) Activation of human and chicken toll-like receptors by Campylobacter spp. Infect Immun 78:1229–1238
Wassenaar TM, Engelskirchen M, Park S, Lastovica A (1997) Differential uptake and killing potential of Campylobacter jejuni by human peripheral monocytes/macrophages. Med Microbiol Immunol 186:139–144
Beery JT, Hugdahl MB, Doyle MP (1988) Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl Environ Microbiol 54:2365–2370
Lee MD, Newell DG (2006) Campylobacter in poultry: filling an ecological niche. Avian Dis 50:1–9
Dhillon AS, Shivaprasad HL, Schaberg D, Wier F, Weber S, Bandli D (2006) Campylobacter jejuni infection in broiler chickens. Avian Dis 50:55–58
Humphrey S, Chaloner G, Kemmett K, Davidson N, Williams N, Kipar A, Humphrey T, Wigley P (2014) Campylobacter jejuni is not merely a commensal in commercial broiler chickens and affects bird welfare. MBio 5:e01364-14
Meade KG, Narciandi F, Cahalane S, Reiman C, Allan B, O’Farrelly C (2009) Comparative in vivo infection models yield insights on early host immune response to Campylobacter in chickens. Immunogenetics 61:101–110
Smith CK, Kaiser P, Rothwell L, Humphrey T, Barrow PA, Jones MA (2005) Campylobacter jejuni-induced cytokine responses in avian cells. Infect Immun 73:2094–2100
Li YP, Ingmer H, Madsen M, Bang DD (2008) Cytokine responses in primary chicken embryo intestinal cells infected with Campylobacter jejuni strains of human and chicken origin and the expression of bacterial virulence-associated genes. BMC Microbiol 8:107
Smith CK, Abuoun M, Cawthraw SA, Humphrey TJ, Rothwell L, Kaiser P, Barrow PA, Jones MA (2008) Campylobacter colonization of the chicken induces a proinflammatory response in mucosal tissues. FEMS Immunol Med Microbiol 54:114–121
Myszewski MA, Stern NJ (1991) Phagocytosis and intracellular killing of Campylobacter jejuni by elicited chicken peritoneal macrophages. Avian Dis 35:750–755
Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668
Bacon DJ, Szymanski CM, Burr DH, Silver RP, Alm RA, Guerry P (2001) A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol Microbiol 40:769–777
Szymanski CM, Michael FS, Jarrell HC, Li J, Gilbert M, Larocque S, Vinogradov E, Brisson JR (2003) Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J Biol Chem 278:24509–24520
McNally DJ, Jarrell HC, Li J, Khieu NH, Vinogradov E, Szymanski CM, Brisson JR (2005) The HS:1 serostrain of Campylobacter jejuni has a complex teichoic acid-like capsular polysaccharide with nonstoichiometric fructofuranose branches and O-methyl phosphoramidate groups. FEBS J 272:4407–4422
McNally DJ, Jarrell HC, Khieu NH, Li J, Vinogradov E, Whitfield DM, Szymanski CM, Brisson JR (2006) The HS:19 serostrain of Campylobacter jejuni has a hyaluronic acid-type capsular polysaccharide with a nonstoichiometric sorbose branch and O-methyl phosphoramidate group. FEBS J 273:3975–3989
van Alphen LB, Wenzel CQ, Richards MR, Fodor C, Ashmus RA, Stahl M, Karlyshev AV, Wren BW, Stintzi A, Miller WG, Lowary TL, Szymanski CM (2014) Biological roles of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. PLoS One 9:e87051
Bachtiar BM, Coloe PJ, Fry BN (2007) Knockout mutagenesis of the kpsE gene of Campylobacter jejuni 81116 and its involvement in bacterium–host interactions. FEMS Immunol Med Microbiol 49:149–154
Jones MA, Marston KL, Woodall CA, Maskell DJ, Linton D, Karlyshev AV, Dorrell N, Wren BW, Barrow PA (2004) Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect Immun 72:3769–3776
Rose A, Kay E, Wren BW, Dallman MJ (2012) The Campylobacter jejuni NCTC11168 capsule prevents excessive cytokine production by dendritic cells. Med Microbiol Immunol 201:137–144
Irvine KM, Andrews MR, Fernandez-Rojo MA, Schroder K, Burns CJ, Su S, Wilks AF, Parton RG, Hume DA, Sweet MJ (2009) Colony-stimulating factor-1 (CSF-1) delivers a proatherogenic signal to human macrophages. J Leukoc Biol 85:278–288
Garcia-Morales C, Nandi S, Zhao D, Sauter KA, Vervelde L, McBride D, Sang HM, Clinton M, Hume DA (2015) Cell-autonomous sex differences in gene expression in chicken bone marrow-derived macrophages. J Immunol 194:2338–2344
McNally DJ, Lamoureux MP, Karlyshev AV, Fiori LM, Li J, Thacker G, Coleman RA, Khieu NH, Wren BW, Brisson JR, Jarrell HC, Szymanski CM (2007) Commonality and biosynthesis of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. J Biol Chem 282:28566–28576
Karlyshev AV, Linton D, Gregson NA, Lastovica AJ, Wren BW (2000) Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol Microbiol 35:529–541
Chintoan-Uta C, Cassady-Cain RL, Al-Haideri H, Watson E, Kelly DJ, Smith DG, Sparks NH, Kaiser P, Stevens MP (2015) Superoxide dismutase SodB is a protective antigen against Campylobacter jejuni colonisation in chickens. Vaccine 33:6206–6211
Kogut MH, Swaggerty C, He H, Pevzner I, Kaiser P (2006) Toll-like receptor agonists stimulate differential functional activation and cytokine and chemokine gene expression in heterophils isolated from chickens with differential innate responses. Microbes Infect 8:1866–1874
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Iovine NM, Pursnani S, Voldman A, Wasserman G, Blaser MJ, Weinrauch Y (2008) Reactive nitrogen species contribute to innate host defense against Campylobacter jejuni. Infect Immun 76:986–993
R Development Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. ISBN 3-900051-07-0, http://www.R-project.org. Accessed 2016
van Mourik A, Steeghs L, van Laar J, Meiring HD, Hamstra HJ, van Putten JP, Wosten MM (2010) Altered linkage of hydroxyacyl chains in lipid A of Campylobacter jejuni reduces TLR4 activation and antimicrobial resistance. J Biol Chem 285:15828–15836
Matsuguchi T, Musikacharoen T, Ogawa T, Yoshikai Y (2000) Gene expressions of Toll-like receptor 2, but not Toll-like receptor 4, is induced by LPS and inflammatory cytokines in mouse macrophages. J Immunol 165:5767–5772
Nomura F, Akashi S, Sakao Y, Sato S, Kawai T, Matsumoto M, Nakanishi K, Kimoto M, Miyake K, Takeda K, Akira S (2000) Cutting edge: endotoxin tolerance in mouse peritoneal macrophages correlates with down-regulation of surface toll-like receptor 4 expression. J Immunol 164:3476–3479
Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS (2005) Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A 102:99–104
Brown SL, Riehl TE, Walker MR, Geske MJ, Doherty JM, Stenson WF, Stappenbeck TS (2007) Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J Clin Investig 117:258–269
Hyun J, Romero L, Riveron R, Flores C, Kanagavelu S, Chung KD, Alonso A, Sotolongo J, Ruiz J, Manukyan A, Chun S, Singh G, Salas P, Targan SR, Fukata M (2015) Human intestinal epithelial cells express interleukin-10 through Toll-like receptor 4-mediated epithelial-macrophage crosstalk. J Innate Immun 7:87–101
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PK and SA initiated and funded this study, and MS and LV supervised and consulted the study direction and progress. AV carried out first two independent experiments with chBMMs and SC prepared all huMDMs. SK carried out all other experiments. SK drafted the manuscript and SC, MS and LV contributed the revision of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We gratefully thank Prof B. Wren for the supply of the C. jejuni strains used in this study, and Dr C. Chinton-Uta and Dr R. Cassady-Cain for comments and technical assistance during the study.
Funding
We acknowledge funding from the Scottish Government via the Strategic Partnership for Animal Science Excellence (SPASE) initiative, and the Scottish Government via the Strategic Partnership for Animal Science Excellence initiative and the Rural & Environmental Science and Analytical Services programme of research for 2016-2012. This work was supported by the Institute Strategic Grant funding from the BBSRC (BB/J004227/1, BB/J004219/1 and BB/P013740/1).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1.
Campylobacter jejuni capsule and MeOPN mutants elicit an enhanced cytokine responses in mBMDCs. The mBMDCs were differentiated from vials of frozen C57BL/6 mouse bone marrow following the protocol of Rose et al. [31] in the presence of GM-CSF (final concentration of 20 ng/mL; Peptro Tech) for 7 days. Live or fixed C. jejuni 11168H (WT), Δcj1417 (MeOPN) and Δcj1439 (capsule) mutants were incubated with mBMDCs at MOI 50 for 3 h. Salmonella LPS was used as a positive control, while PBS in the medium was used as a negative control. Graphs show representative results of two independent experiments. Error bars are the standard deviation of two independent experiments. Asterisks indicate a statistically significant difference of mRNA levels of inflammatory-related cytokines compared to C. jejuni WT (*P < 0.05; **P < 0.01).
Additional file 2.
Analysis of cytokine responses of chBMMs when inoculated at an MOI 100 with the C. jejuni WT and capsule or MeOPN deficient strains. Live or fixed C. jejuni 11168H (WT), Δcj1417 (MeOPN) and Δcj1439 (capsule) mutants were incubated with chBMMs at MOI 100 for 3 and 6 h. Salmonella LPS (100 ng/mL) was used as a positive control and PBS in the medium as a negative control. Graphs show representative results of three independent experiments. Error bars are the standard deviation of three independent experiments. Asterisks indicate a statistically significant difference of mRNA levels of inflammation-related cytokines compared to PBS in the medium (*P < 0.01; **P < 0.001).
Additional file 3.
Analysis of TLR4 transcription in chBMMs stimulated wild-type C. jejuni and capsule or MeOPN mutants. Live or fixed C. jejuni 11168H (WT), Δcj1417 (MeOPN) and Δcj1439 (capsule) mutants were incubated with chBMMs at MOI 50 for 3 h. Salmonella LPS (100 ng/mL) was used as a positive control and PBS in the medium as a negative control. Graphs show representative results of three independent experiments, and error bars are the standard deviation of three independent experiments. Asterisks indicate a statistically significant difference of TLR4 transcripts compared to PBS in the medium (P < 0.05).
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kim, S., Vela, A., Clohisey, S.M. et al. Host-specific differences in the response of cultured macrophages to Campylobacter jejuni capsule and O-methyl phosphoramidate mutants. Vet Res 49, 3 (2018). https://doi.org/10.1186/s13567-017-0501-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-017-0501-y