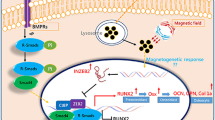
Abstract
Background
Multiple strategies have been proposed to promote the differentiation potential of mesenchymal stem cells (MSCs), which is the fundamental property in tissue formation and regeneration. However, these strategies are relatively inefficient that limit the application. In this study, we reported a novel and efficient strategy, nanosecond pulsed electric fields (nsPEFs) stimulation, which can enhance the trilineage differentiation potential of MSCs, and further explained the mechanism behind.
Methods
We used histological staining to screen out the nsPEFs parameters that promoted the trilineage differentiation potential of MSCs, and further proved the effect of nsPEFs by detecting the functional genes. In order to explore the corresponding mechanism, we examined the expression of pluripotency genes and the methylation status of their promoters. Finally, we targeted the DNA methyltransferase which was affected by nsPEFs.
Results
The trilineage differentiation of bone marrow-derived MSCs was significantly enhanced in vitro by simply pre-treating with 5 pulses of nsPEFs stimulation (energy levels as 10 ns, 20 kV/cm; 100 ns, 10 kV/cm), due to that the nsPEFs demethylated the promoters of stem cell pluripotency genes OCT4 and NANOG through instantaneous downregulation of DNA methylation transferase 1 (DNMT1), thereby increasing the expression of OCT4 and NANOG for up to 3 days, and created a treatment window period of stem cells.
Conclusions
In summary, nsPEFs can enhance MSCs differentiation via the epigenetic regulation and could be a safe and effective strategy for future stem cell application.
Similar content being viewed by others
Introduction
Mesenchymal stem cells (MSCs) have been used for cell-based therapies due to their significant contribution in tissue development and regeneration [1]. Two major characteristics of MSCs, i.e., the self-renewal capacity and the differentiation potential, empower the clinical application by enlarging the cell population and contributing to the on-site neo-tissue formation. For example, bone marrow-derived MSCs, which can differentiate into osteo-, adipo-, and chondro-lineages [2], have brought positive clinical results in treating bone and cartilage defects [3, 4]. The differentiation potential of MSCs, both in vitro and in vivo, directly related to the therapeutic efficacy, depends on the tissue source (e.g., tissue derivation, health status of donor site), the cell isolation, and culture conditions [5,6,7]. In order to maintain the therapeutic characteristics of MSCs, many attempts have been tried, such as treatments of growth factors [8, 9] or preconditioned with hypoxia [10]. However, these methods are functioning below an effective threshold that limit the application, thus more advanced methods are needed.
Pulsed electrical stimulation (PES) has been proven to have multiple biological effects on cells for the short-term permeabilization, which mostly depends on the parameters of electric field strength (from millivolt/cm, mV/cm, to megavolt/cm, MV/cm) and stimulation duration (from nanosecond, ns, to second, s) [11]. PES-based technologies have been applied for tumor therapy, because it can induce cell apoptosis at the range of kV-MV/cm within ns-μs stimulation duration, and have been also used for electroporation for gene delivery at the range of V/cm, μs-s [11]. Nanosecond pulsed electric fields (nsPEFs), a novel technology with relatively short duration (nanoseconds, ns) and subsequent high voltages (up to kV/cm), are emerging in cell researches and have been reported to have the modulation effects on stem cells. nsPEFs can incur more comprehensive biological effects, compared with traditional pulsed electric fields (PEFs) which are above millisecond or microsecond. As the duration is shorter than the charging time of cell membrane, nsPEFs can further affect intracellular structures [11]. The biological responses of nsPEFs have been previously reported in algae cells [12], and in human cancer cell lines with the mobilization of intracellular Ca2+ and activation of signaling pathways [13, 14]. Besides, study has shown that nsPEFs (80 ns, 20 kV/cm, 1 Hz) may induce demethylation and activation of Suppressors of cytokine signaling (Socs) in Hela S3 cells [15]. The comprehensive and individualized reactions of cells are based on varied combinations of parameters (duration, voltage, frequency, and number of pulses) of nsPEFs, and the physical and biological properties of cells [13, 16]. nsPEFs have been studied as a possible therapeutic intervention for cancer [17,18,19], but little is known about their effects on regulating cell phenotype and differentiation of stem cells.
Previously, our group has found that nsPEFs could affect chondrocyte phenotype through regulating Wnt/β-catenin signaling pathway [20]. Recently, we found that nsPEFs at the levels of 10 ns at 20 kV/cm, 60 ns at 5 kV/cm, 60 ns at 10 kV/cm, 60 ns at 20 kV/cm, and 100 ns at 10 kV/cm, separately, could upregulate chondrogenic gene expression of MSCs [21]. Notably, cells can respond to physical energy epigenetically [22]; therefore, it is possible that nsPEFs have a role in epigenetic regulation of MSCs. In this study, we find that nsPEFs with specific parameters can make MSCs more susceptible to induced differentiation. In addition, we reveal that nsPEFs can downregulate DNA methylation transferase 1 (DNMT1) temporarily, and switch on the negative feedback loop between DNMT1 and OCT4/NANOG. In contrast, overexpression of DNMT1 can block the effect of nsPEFs.
Methods
Harvest and culture of mesenchymal stem cells
Porcine bone marrow mesenchymal stem cells (pMSCs) were harvested from three Guizhou mini-pigs (Peking University, Laboratory Animal Center, PKU-LAC) at 6 to 10 months old (approved by IACUC, PKU-LAC). Femur and tibia of the pigs were drilled before bone marrow was washed out with phosphate-buffered saline (PBS) and collected in 50 ml centrifuge tubes. Human bone marrow mesenchymal stem cells (hMSCs) were obtained from the bone marrow of three patients receiving total hip arthroplasty (male, 62 years old; male, 79 years old; female, 82 years old, from the People’s Hospital, Beijing, with IRB approval) and collected in 50 ml centrifuge tubes. After centrifuged at 1000 rpm and supernatant removed, the collected cells were suspended with medium containing 90% Dulbecco’s modified Eagle’s medium (DMEM, Gibco), 10% fetal bovine serum (FBS, Gibco), and 1% penicillin/streptomycin (PS, Amresco) and cultured at 37 °C in humidified atmosphere with 5% CO2. The cultured medium was changed every 3 days until the cells reached 85% of confluency. Then, they were trypsinized with 0.25% trypsin (27250–018, Invitrogen), and MSCs at passage 5 were used for all subsequent experiments.
Application of nsPEFs
One million of MSCs were suspended in 1 mL of culture medium in gap cuvette (BTX, catalog number 45-0125, 45-0126), and were subjected to 5 pulses of nsPEFs (10 ns at 20 kV/cm, 60 ns at 5 kV/cm, 60 ns at 10 kV/cm, 60 ns at 20 kV/cm, and 100 ns at 10 kV/cm, 1 Hz). And the time interval between two pulses is 1 s [21]. MSCs not subjected to nsPEFs served as control. The nsPEFs generator was applied as previously described [23]. The voltage waveform was monitored by a Digital phosphor oscilloscope (DPO4054, Tektronix) with a probe (P6015A, Tektronix).
qRT-PCR
Total RNA was extracted and isolated from MSCs in each stimulation condition with Trizol Reagent (New Industry) following the standard protocol and quantified with Nanodrop spectrophotometer (ND-1000, Thermo). Then, the reverse transcription reaction was performed on 500 ng of RNA with M-MLV reverse transcriptase (C28025, Sigma) and oligo (dT) (FSK-201, TOYOBO) in a PCR thermal. Quantitative real-time polymerase chain reactions (qRT-PCR) were performed in the PCR system (Pikoreal 96, Thermo) with RealMasterMix SYBR Green (FP202, Tiangen) following the manufacturer’s procedures. The expression levels of stem cell pluripotency genes and trilineage differentiation-related genes were analyzed. The primers were listed in Supplementary Table 1. The target genes of each sample were normalized to the values of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as internal control. Relative expression of each gene was expressed as fold changes by the 2−ΔΔCt method. The experiment was repeated three times, with five technological repeats for each assay.
Histology analysis of osteo-, adipo-, and chondrogenic differentiation
The treated MSCs were cultured with osteogenic induction medium, adipogenic induction medium, and chondrogenic induction medium [24] at 37 °C in humidified atmosphere with 5% CO2, respectively. Media was changed every 3 days. After 14 days, cell cultures were stained with Alizarin Red (AR, for osteogenic induction) and Oil Red O (ORO, for adipogenic induction) and Alcian Blue (AB, for chondrogenic induction) staining, respectively, and followed by extraction and measurement of O.D. values of AR staining at 550 nm, ORO staining at 510 nm, and AB staining at 620 nm. The experiment was repeated three times, with five technological repeats for each assay.
Western blotting
MSCs were lysed by RIPA lysis buffer (R0020, Solarbio) with fresh protease inhibitor of 0.1% phenylmethanesulfonyl fluoride 2 h after nsPEFs, and mixed with 4× SDS loading buffer (P1015, Solarbio). The western blotting was carried out according to the manufacturer’s protocols. Rabbit polyclonal antibodies against DNMT1 (24206-1-AP, Proteintech), DNMT3a (3598, Cell Signaling), DNMT3b (orb372330, Biorbyt), and β-actin (4970S, Cell Signaling) were used in combination with secondary HRP-linked antibody of anti-rabbit IgG (7074S, Cell Signaling). The complex of the antigen and the antibody was detected with TANON 1600 Gel Imaging System (Tanon Science&Technology Co., Ltd., Shanghai), and the expression level of protein is analyzed with Tanon Gis (Tanon Science&Technology Co., Ltd., Shanghai).
Overexpressing of DNMT1 in MSCs
For tet-on DNMT1 systems, we synthesized the coding sequence of pDNMT1 gene from GENEWIZ by chemical method. The amplified sequence pDNMT1 was then cloned into a pFU-tetO lentivirus backbone (19778, Addgene) linearizing with EcoR1 restriction enzyme. The FUdeltaGW-rtTA (19780) and third-generation lentiviral helper plasmid (12253, 12252, 12251) were purchased from Addgene. pFU-tetO-pDNMT1 and FUdeltaGW-rtTA were co-transfected into MSCs. Plasmids with GFP genes were used as control. Because there was almost no significant differences between nsPEFs with the two set parameters (10 ns at 20 kV/cm, and 100 ns at 10 kV/cm), nsPEFs of 100 ns at 10 kV/cm was used for studying the effects of downregulation of DNMT1. After stimulation by nsPEFs, doxycycline (Dox) was added to MSCs at 1 μg/ml for 2 h. The expression levels of GFP and DNMT1 were evaluated by western blotting. The primers and annealing temperatures used for PCR of GFP and DNMT1 are listed in Supplementary Table 3. The experiment was repeated three times, with five technological repeats for each assay.
Statistical analysis
Results were presented as the mean ± SD/SEM, and was normalized to the control group defined as One-way ANOVA was carried out with the least significant difference (LSD) using Prism 5.03 software (GraphPad), depending on the group numbers. The statistical significance level was set as p < 0.05.
Results
Pre-conditioning with nsPEFs enhances trilineage differentiation potential of pMSCs
Stem cell properties of MSCs can be assessed by assaying the potential to differentiate along the osteogenic, adipogenic, and chondrogenic lineages [25]. In order to optimize the treatment conditions of nsPEFs, five sets of nsPEFs parameters (i.e., 10 ns at 20 kV/cm, 60 ns at 5 kV/cm, 60 ns at 10 kV/cm, 60 ns at 20 kV/cm, and 100 ns at 10 kV/cm) were firstly screened by the differentiation assays of pMSCs (Fig. 1a). We found that only nsPEFs of 10 ns at 20 kV/cm and 100 ns at 10 kV/cm had the effect of enhancing trilineage differentiation potential of pMSCs, the other three sets of parameters just enhanced one type of differentiation ability (Table 1, Fig. 1b, c).
A single nsPEF treatment (5 pulses, less than 10 s) can enhance the differentiation of MSCs. a Schematic of MSCs stimulated by nsPEFs. One million of MSCs were suspended in culture medium in gap cuvette and were subjected to 5 pulses of nsPEFs (i.e., 10 ns at 20 kV/cm and 100 ns at 10 kV/cm). And the time interval between two pulses is 1 s. Then, the trilineage differentiation induction was carried out. b Alizarin Red S, Oil red O staining, and Alcian blue staining for osteogenic, adipogenic, and chondrogenic differentiation at day 14, insets show the no-staining counterparts. c Quantification of differentiation into osteogenic, adipogenic, and chondrogenic lineages. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS, p > 0.05) d–f qRT-PCR for the mRNA levels of genes associated with trilineage differentiation (osteogenic: RUNX2, OCN; adipogenic: PPARγ, LPL; chondrogenic: SOX9, COLII) respectively at day 14. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS, p > 0.05)
The expression levels of differentiation genes were also evaluated at day 14. Osteogenic transcription factor RUNX2 was upregulated by 10.53 ± 1.91- and 9.03 ± 1.77-fold by nsPEFs (10 ns at 20 kV/cm, and 100 ns at 10 kV/cm) (Fig. 1d), main regulating valves for adipogenic differentiation PPARγ was improved by 6.06 ± 0.78-fold (10 ns at 20 kV/cm) and 9.93 ± 1.42-fold (100 ns at 10 kV/cm) (Fig. 1e), chondrogenic transcription factor SOX9 was increased by 10.50 ± 1.95-fold (10 ns at 20 kV/cm) and 10.82 ± 1.09-fold (100 ns at 10 kV/cm) (Fig. 1f). The expressions of other related functional genes (OCN, ALP; LPL, AP2; COLII, AGG) can be upregulated for 5–10-folds compared to the control group (Fig. 1d–f and Fig. S1A-C). Taken together, these data suggest that the biological effects of nsPEFs depend on the time and energy levels of treatment. Only two sets of parameters, i.e., 10 ns at 20 kV/cm, and 100 ns at 10 kV/cm, could enhance the differentiation potential of pMSCs.
Optimized nsPEFs do not influence the proliferation of pMSCs
Proliferation of pMSCs was evaluated with MTT assay over 7 days after preconditioning with nsPEFs, and nsPEF treatments did not influence the proliferation of pMSCs (Fig. S2A). Moreover, cell cycle analysis and colony-forming units (CFU) assays were performed to evaluate the effects of nsPEFs. There were no significant differences in cell cycles (Fig. S2B) or CFU numbers (Fig. S2C) between nsPEF treatments and control groups. These data indicate that our optimized nsPEFs parameters do not influence the clonogenicity and proliferation of MSCs.
nsPEFs enhance gene expressions of OCT4 and NANOG via removing the methylation of their promoters
OCT4 and NANOG are critical transcriptional factors for stem cell pluripotency [26]. To further explore the cellular molecular mechanisms of the biological effects caused by nsPEFs, the expression levels of pluripotency genes OCT4 and NANOG were examined. Interestingly, an instant elevation of OCT4 and NANOG was found after 2 h of nsPEF treatment both in porcine MSCs (pMSCs) and human MSCs (hMSCs) (Fig. 2a). The expression of OCT4 increased significantly with 2.89 ± 0.30-fold changes in pMSCs (p = 0.0029), and 4.82 ± 0.97-fold in hMSCs (p = 0.0044) for 10 ns at 20 kV/cm nsPEF treatments; 3.56 ± 0.30-fold in pMSCs (p = 0.001), and 3.42 ± 0.86-fold in hMSCs (p = 0.0476) for 100 ns at 10 kV/cm of nsPEF treatments (Fig. 2a). The expression of NANOG was also upregulated significantly (pMSCs 1.68 ± 0.27-fold, p = 0.0396 and 1.7 ± 0.16-fold, p = 0.0044; hMSCs 2.44 ± 0.15-fold, p = 0.0005 and 1.96 ± 0.21-fold, p = 0.0093) in both nsPEF treatment groups (10 ns at 20 kV/cm, and 100 ns at 10 kV/cm) (Fig. 2a). We then tracked the gene expression levels of OCT4 and NANOG of pMSCs at 3 days and 7 days after nsPEFs preconditioning and found that the upregulated OCT4 subsequently decreased over 7 days (Fig. S3A and C), while the expression levels of NANOG remained the same after nsPEFs (Fig. S3B and D). In addition to the gene expression levels of OCT4 and NANOG, we further examined the epigenetic modification by using bisulfite sequencing analysis. With the precondition of nsPEFs, a clearly drop was found in the methylation levels of CpG sites of OCT4 and NANOG promoters, compared with non-treated pMSCs control group (Fig. 2b, c). Therefore, these data suggest that nsPEFs can directly function on MSCs by demethylating the promoter region of OCT4 and NANOG.
nsPEFs promote OCT4 and NANOG expressions with increasing demethylation level of promoter. a qRT-PCR for the expressions of OCT4 and NANOG of pMSCs and hMSCs at 2 h after stimulation by nsPEFs. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA. *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS, p > 0.05) b Bisulfite sequencing analysis of OCT4 and NANOG promoter of pMSCs at 2 h after stimulation by nsPEFs. Each CpG is represented by a circle in the 50–30 orientation; each row represents the methylation state of each CpG in one bacterial clone of PCR product. White circle indicates unmethylated CpG; black circle indicates methylated CpG. c Percentage of CpG demethylation for each promoter. (Values are mean ± SD, n = 3, one-way ANOVA, *p = 0.0329, *p = 0.0171)
To further investigate if the instant upregulation of pluripotency genes was a universal effect for all stem cell types, we also evaluated the OCT4 and NANOG changes in human embryonic stem cells (hESCs, details are in supplementary documents) at 2 h after nsPEFs preconditioning. Interestingly, we found that only nsPEFs with parameter of 100 ns at 10 kV/cm can enhance the gene expressions of OCT4 (4.92 ± 1.00-fold, p = 0.0097) and NANOG (4.63 ± 1.16-fold, p = 0.0223) of hESCs significantly, but not with 10 ns at 20 kV/cm (Fig. S3E and F).
nsPEFs temporally decrease DNMT1 expression
We next aimed to gain insights into how the hypomethylation of the OCT4 and NANOG promoters was regulated by nsPEFs. DNA methylation of CpG dinucleotides is catalyzed by at least three different DNA methylation transferases (DNMTs), including DNMT1, DNMT3a, and DNMT3b. And DNMT3a and DNMT3b function primarily as de novo methyltransferases that establish DNA methylation patterns, while DNMT1 is a key enzyme that maintains methylation patterns following DNA replication [27]. The DNMTs are essential for maintaining the methylation pattern in stem cells and for regulating their self-renewal and differentiation [24, 28]. The protein expression of DNMT1 substantially dropped by 0.58 ± 0.11- and 0.27 ± 0.05-fold respectively at 2 h after nsPEF treatment (10 ns at 20 kV/cm; 100 ns at 10 kV/cm) in pMSCs, while declined to 0.69 ± 0.02- and 0.56 ± 0.06-fold in hMSCs (Fig. 3a). Gene expression of DNMT1 decreased significantly to 0.3 ± 0.07- and 0.3 ± 0.06-fold in pMSCs, and to 0.52 ± 0.03- and 0.41 ± 0.06-fold in hMSCs (Fig. 3b). However, the levels of DNMT3a and DNMT3b did not change in both pMSCs and hMSCs (Fig. S4A and B). To confirm the function of elevated DNMT1, the 5-methylcytosine contents which reflect global DNA methylation level were measured at 2 h after nsPEFs. The global DNA methylation analysis revealed a 0.39 ± 0.06- or 0.51 ± 0.05-fold expression in nsPEFs-preconditioned groups compared with control group (Fig. 3c).
DNMT1 responds to nsPEFs with a short window of 3 days. a Western blot for DNMT1 protein expression level of pMSCs and hMSCs at 2 h after stimulation by nsPEFs. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS, p > 0.05) b qRT-PCR for the expression of DNMT1 of pMSCs and hMSCs at 2 h after stimulation by nsPEFs. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS, p > 0.05) c The global DNA methylation level at 2 h after stimulation by nsPEFs. (n = 6, one-way ANOVA, ****p < 0.0001, NS = 0.1515, ***p = 0.0002) d Protein quantification for the expression of DNMT1 at 2, 12, 24, and 72 h after stimulation by nsPEFs. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS, p > 0.05)
To investigate how long the effects can last, protein expression levels of DNMT1 in pMSCs at 2, 12, 24, and 72 h after nsPEFs were examined. After nsPEF treatment, the expression of DNMT1 gradually increased from a lower level at 2 h, and peaked at 24 h, which was greatly higher than control groups, and then entered the end point of a dynamic equilibrium to the levels of control groups at 72 h (Fig. 3d).
Overexpression of DNMT1 blocks the upregulation of OCT4 and NANOG induced by nsPEFs
To further justify if nsPEFs-reduced DNMT1 directly affected the expressions of OCT4 and NANOG, as well as the subsequent differentiation of pMSCs, we established a tet-on system to drive the DNMT1 expression in pMSCs (GFP as system control) (Fig. 4a). There were minor differences between the two sets of nsPEFs parameters (10 ns at 20 kV/cm vs. 100 ns at 10 kV/cm) in terms of the biological effects in earlier experiments. However, nsPEFs with the two parameters led to a very similar trend of regulation on DNMT1, trilineage differentiation as well as pluripotency genes of MSCs. Based on it, we assumed that nsPEFs with these two parameters regulated the cells under same or similar mechanism. And nsPEFs at the levels of 100 ns at 10 kV/cm were used in this section. Overexpression of DNMT1 by the tet-on system increased the protein expression of DNMT1 by 1.33 ± 0.09-fold (p = 0.0138), which indicated that we successfully established the DNMT1 overexpression model. Treatment of nsPEFs lowered the protein expression of DNMT1 by 0.34 ± 0.06-fold in GFP control group (Fig. 4b), which matched with the earlier results in pMSCs and hMSCs (Fig. 3a). Notably, the enhanced expression levels of OCT4 (3.50 ± 0.77-fold, p = 0.0309, nsPEFs+ group) and NANOG (1.95 ± 0.22-fold, p = 0.0121, GFP+/nsPEFs+ group) were blocked by overexpression of DNMT1, and the expressions of OCT4 and NANOG stayed unchanged at 2 h after nsPEF treatment (Fig. 4c, DNMT1+/nsPEFs+ group). We then evaluated the percentage of CpG demethylation of OCT4 and NANOG promoters with bisulfite sequencing analysis in this DNMT1 overexpression model (Fig. 4d), and the results were consistent with the genes expression levels (Fig. 4c). Taken together, these data show that overexpression of DNMT1 can block the effects of nsPEFs on gene expressions of OCT4 and NANOG in pMSCs.
Overexpression of DNMT1 blocks demethylation caused by nsPEFs. a Schematic of MSCs stimulated by nsPEFs with overexpression of DNMT1. b Western blot for DNMT1 after pre-treated with nsPEFs with overexpression of GFP or DNMT1. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS, p > 0.05) c Quantitative RT-PCR for the expressions of OCT4 and NANOG at 2 h after stimulation by nsPEFs. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS, p > 0.05) d Percentage of CpG demethylation for OCT4 and NANOG promoter. (Values are mean ± SD, n = 3, one-way ANOVA, *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS, p > 0.05)
Overexpression of DNMT1 blocks the subsequent effects of nsPEFs on stem cell differentiation
To further investigate if DNMT1 overexpression erased the subsequent differentiation performance of MSCs enhanced by nsPEFs, both trilineage differentiation and related functional genes were evaluated (Fig. 5). Osteogenic differentiation, which was indicated by the quantification of alizarin red staining intensity (Fig. 5a, b), was increased by 1.37 ± 0.09-fold (p = 0.0071, GFP+/nsPEFs+ group) by nsPEFs (100 ns, 10 kV/cm), and decreased by 0.78 ± 0.06-fold by overexpression of DNMT1 (p = 0.0068, DNMT1+/nsPEFs− group). Meanwhile, there was no significant difference between control group (GFP+/nsPEFs− group) and nsPEFs stimulated DNMT1 overexpression group (p = 0.4912, DNMT1+/nsPEFs+ group). The differentiation performance of pMSCs into adipogenic lineage (oil-red O staining) and chondrogenic lineage (Alcian blue staining) shared the same trends as osteogenic differentiation (Fig. 5a, b). The expression levels of trilineage differentiation-related key genes (osteogenic: RUNX2, OCN; adipogenic: PPARγ, LPL; chondrogenic: SOX9, COLII) showed similar trends with the differentiation assays, that all functional genes were upregulated in GFP+/nsPEFs+ groups and had no significant change in DNMT1+/nsPEFs+ groups (Fig. 5c).
Overexpression of DNMT1 hinders the differentiation potential of MSCs caused by nsPEFs. a Alizarin Red S, Oil red O staining, and Alcian blue staining for osteogenic differentiation, adipogenic and chondrogenic differentiation at day 14; insets show the no-staining counterparts. b Quantification of differentiation into osteogenic, adipogenic, and chondrogenic lineages. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, NS, p > 0.05) c MSCs were induced to undergo osteogenic, adipogenic, and chondrogenic differentiation at day 14, and qRT-PCR was performed. Trilineage differentiation-related key genes, osteogenic: RUNX2, OCN; adipogenic: PPARγ, LPL; chondrogenic: SOX9, COLII (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, N, p > 0.05)
Discussion
In this study, we discovered that a simple precondition of MSCs with nsPEFs can enhance the differentiation potential of cultured stem cells. We then investigated the cellular and molecular mechanism of this phenomenon and found that nsPEFs can remove the methylation of promoters of the pluripotency genes OCT4 and NANOG temporally via downregulating the DNA methyltransferases, in particularly, the DNMT1, where the higher expressions of OCT4 and NANOG were seen (Fig. 6). These nsPEF-induced epigenetic responses probably can further establish a hypersensitive phase for cell differentiation and thus performed better in all trilineage differentiation assays.
Schematic illustration of the possible molecular mechanisms induced by nsPEFs in MSCs. nsPEFs stimulation (5 pulses of electrical stimulation at levels of 10 ns at 20 kV/cm, or 100 ns at 10 kV/cm; the time interval between two pulses is 1 s) could enhance trilineage differentiation of both MSCs. In terms of the mechanism, nsPEFs could downregulate DNMT1, temporally unlock the stabilizer of DNA methylation, and lead to the elevated stem cell pluripotency gene expression of OCT4 and NANOG
Early mechanistic studies on the biological effects of nsPEFs have indicated that the short-term high-energy stimulation can influence the intracellular membranes by electroporation and permeabilization [29], while their effects on epigenetic regulation are rarely reported. Treatment of nsPEFs with sufficient short-pulse durations and rapid rise times can induce supra-electroporation in all membranes of a cell, and extensively penetrate all membranes of organellars, supported by patch-clamp and fluorescent imaging results [29]. We here further explored the biological effects of nsPEFs and found that nsPEFs can regulate DNA modification and gene expression epigenetically.
Notably, the biological effects on cells depend on the energy level and duration of nsPEFs. As the durations decrease, the effects of the electric fields on cells shift from the cell membrane to the organelle membrane. Relatively short durations and high field strengths (hundreds kV/cm) can potentially affect the intracellular membrane while restricting total energy and narrowing down the previously broad biological effects [11]. As the two electrical parameters of nsPEFs used (10 ns at 20 kV/cm and 100 ns at 10 kV/cm) have similar voltages, it is reasonable to assume that they incur similar biological effects on MSCs. Study has shown that higher energy and longer duration (300 ns, 1.8 kV/cm) can cause cell apoptosis [30]. Indeed, it raises the safety concerns in the further application in stem cells. Our group has narrowed down a safety and effective range of nsPEFs for studying the biological effects of nsPEFs on MSCs [21]. In our previous study, we have checked the cell viability of MSCs by flow cytometry after 1 h of nsPEFs stimulation at the levels of 10 ns at 20 kV/cm and 100 ns at 10 kV/cm, which showed no difference with regular cultured cells [21]. In addition, the current study also showed that the cell proliferation, cell cycle, and colony-forming capacity of MSCs are not affected by nsPEFs at the defined levels. Our results indicated that these two sets of nsPEFs parameter (10 ns at 20 kV/cm; 100 ns at 10 kV/cm) are safe for MSCs.
Here, we found that nsPEFs can enhance stem cell differentiation through temporally fine-tuning gene expressions of OCT4 and NANOG. OCT4, SOX2, and NANOG comprise a core transcriptional network that regulates self-renewal and pluripotency of stem cells, and are key elements for somatic cell to reprogram into iPSCs [31, 32]. These pluripotency genes are related to the differentiation potential of MSCs and can be seen as early-stage indicators and regulators of stem cell potencies [33, 34]. Many biophysical approaches can regulate the expressions of these genes and the differentiation abilities of stem cells. For instance, low-intensity pulsed ultrasound stimulation could upregulate NANOG expression and the subsequent osteogenic differentiation of MSCs [35]; continuous hypoxia (1% oxygen concentration) has been used to enhance and accelerate the osteogenic ability of MSCs with the upregulation of OCT4 [36]; Overexpression of pluripotency genes can promote the differentiation of MSCs [33, 37]. In the study, we reported a similar effect of nsPEFs in regulating stem cell behaviors, that nsPEFs (10 ns at 20 kV/cm; 100 ns at 10 kV/cm) can efficiently upregulate OCT4 and NANOG for 2–4-folds in human and porcine MSCs, and the precondition with 5 pulses can be done in 10 s. Our nsPEF treatment method provides a simpler and more effective way in regulating stem cells with the similar effect compared with the reported physical, chemical, or biotechnological methods, most of which request full-time consuming and complicated operation procedures.
We also found a feedback regulation between DNMT1 and OCT4, NANOG in MSCs. Previous studies have proposed that partial DNA demethylation in the OCT4 and NANOG promoter regions are required for gene activation in ESCs, iPSCs, and other cell types [38,39,40,41]. OCT4 and NANOG are hypomethylated in human ESCs and induced pluripotent stem cells (iPSCs) but are hypermethylated in their fibroblast derivatives [42]. DNMT1 has been shown to contribute to the methylation of OCT4 and NANOG during mouse embryonic cell differentiation in vivo [43]. These research results have indicated that DNMT1 plays an important role in tissue development, and it can block the expressions of pluripotency genes and maintain a fully-differentiated stage of cells after embryo development and terminal differentiation. Here, we found that DNMT1 protein in MSCs was immediately downregulated by nsPEFs, meanwhile OCT4 and NANOG gene expressions were significantly upregulated with demethylation of their promoters. These results suggested that MSCs firstly respond to nsPEFs epigenetically and genetically, and then reconstruct to a hypersensitive phase for differentiation. In addition, when DNMT1 was overexpressed, OCT4 and NANOG genes remained low and unchanged, and this suggested that there was a threshold of DNMT1 in regulating OCT4 and NANOG gene expressions, and certain level of DNMT1 was enough to keep the gate for OCT4 and NANOG in MSCs. We also illustrated that the effects of nsPEFs on the expression of DNMT1 were a dynamic equilibrium procedure, that DNMT1 dropped at 2 h, and gradually increased and peaked to ~ 2-folds at 24 h compared to untreated control, and back to the levels of control groups at 72 h. This phenomena probably can be explained by the regulating effects of OCT4 and NANOG on DNMT1, which has been reported that OCT4 and NANOG can directly bind to the promoter region of DNMT1 to promote the DNMT1 expression in MSCs and fibroblasts [24, 44]. Therefore, together with our findings, we believe that there should be a feedback loop between DNMT1 and OCT4/NANOG in MSCs, and the regulation effect of nsPEFs on MSCs can last for 3 days. On the other hand, unaltered cell proliferation could be attributed to the balanced results of upregulated gene expression of OCT4/NANOG and downregulated DNMT1, for the downregulation of DNMT1 inhibits proliferation [45], and the upregulation of OCT4/NANOG promotes proliferation [26].
nsPEFs can downregulate DNMT1 temporally and enhance gene expressions of OCT4 and NANOG, as well as subsequent osteo-, chondro- and adipo-genetic differentiation of MSCs, which provides us a novel and precise tool for future stem cell research. nsPEF-induced demethylation of the promoter regions of specific genes is able to achieve reversible epigenetic regulation within a treating window of 3 days. Therefore, nsPEFs-based technologies have the potential to be applied in iPSCs research to enhance the yield rate of iPSCs during reprogramming, as both the inhibition of DNMT1 [46,47,48] and electromagnetic fields [22] have been used to improve reprogramming efficiency, as reported previously. Despite all this, the parameters applying in iPSCs could be different from those in MSCs, because we found that only one set paraments of nsPEFs, 100 ns at 10 kV/cm, can regulate OCT4/NANOG in hESCs, which suggested that different cell types may need more detailed parameter segmentations. Another potential future application scenario of nsPEFs is in disease treatment, since the downregulation of DNMT1 has been reported to be able to promote the relief of the osteoarthritic symptoms in chondrocyte [49]. We have analyzed the proteomics of MSCs and found that among 3808 proteins, 59 were increased (fold change > 1.33), and 44 were downregulated (fold change < 0.75) at 2 h after nsPEF treatment, among which 6 proteins were related to epigenetic regulation (data not shown). This result suggests that more genes can be regulated via nsPEFs epigenetically. Given the unlimited parameter combinations, nsPEFs could identify multiple epigenetic targets, and regulate them either temporally or persistently, which have great potential in many bio-applications, such as in development, aging, and regeneration.
Conclusions
This study demonstrates that nsPEFs (as levels in 10 ns at 20 kV/cm, and 100 ns at 10 kV/cm) can enhance differentiation potential of both human and porcine mesenchymal stem cells. As to the molecular mechanism, nsPEFs could temporally unlock the stabilizer of DNA methylation with downregulation of DNMT1, which lead to the upregulation of OCT4 and NANOG. Taken together, nsPEFs preconditioning provides a simple and effective method to improve the differentiation potential of MSCs. And we propose that nsPEFs can further establish a hypersensitive phase for cell differentiation.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AGG:
-
Aggrecan
- ALP:
-
Alkaline phosphatase
- COLII:
-
Type II collagen
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- DNMT:
-
DNA methyltransferase
- Dox:
-
Doxycycline
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- LPL:
-
Lipoprotein lipase
- MSCs:
-
Mesenchymal stem cells
- MTT:
-
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide, Thiazolyl Blue Tetrazolium Bromide
- nsPEFs:
-
Nanosecond pulsed electric fields
- OCN:
-
Osteocalcin
- OCT4:
-
Octamer-binding transcription factor 4
- PPARγ:
-
Peroxisome proliferators-activated receptor-γ
- RUNX2:
-
Runt-related transcription factor 2
- Socs:
-
Suppressors of cytokine signaling
- SOX9:
-
SRY-related high mobility group-box gene9
References
Squillaro T, Peluso G, Galderisi U. Clinical trials with mesenchymal stem cells: an update. Cell Transplant. 2015;25(5):829–48.
Yan L, Munzer SR, Richardson JL. What is stemness? Stud Hist Philos Sci Biol Biomed Sci. 2009;40(4):312–20.
Shinya Y, Hisashi M, Maki I, et al. Cartilage repair with autologous bone marrow mesenchymal stem cell transplantation: review of preclinical and clinical studies. Cartilage. 2014;5(4):196–202.
Hashimoto Y, Nishida Y, Takahashi S, et al. Transplantation of autologous bone marrow-derived mesenchymal stem cells under arthroscopic surgery with microfracture versus microfracture alone for articular cartilage lesions in the knee: a multicenter prospective randomized control clinical trial. Regenerative Ther. 2019;11:106–13.
Erin C, Fei G, Maosong Q, et al. Differential efficacy of human mesenchymal stem cells based on source of origin. J Immunol. 2014;193(9):4381–90.
Tuli R, Seghatoleslami MR, Tuli S, et al. A simple, high-yield method for obtaining multipotential mesenchymal progenitor cells from trabecular bone. Mol Biotechnol. 2003;23(1):37–49.
Jiang Y, Cai Y, Zhang W, et al. Human cartilage-derived progenitor cells from committed chondrocytes for efficient cartilage repair and regeneration. Stem Cells Transl Med. 2016;5(6):733–44.
Sotiropoulou PA, Perez SA, Maria S, et al. Characterization of the optimal culture conditions for clinical scale production of human mesenchymal stem cells. Stem Cells. 2010;24(2):462–71.
Li C, Wu X, Tong J, et al. Comparative analysis of human mesenchymal stem cells from bone marrow and adipose tissue under xeno-free conditions for cell therapy. Stem Cell Res Ther. 2015;6(1):1–13.
Antebi B, Rodriguez LA, Walker KP, et al. Short-term physiological hypoxia potentiates the therapeutic function of mesenchymal stem cells. Stem Cell Res Ther. 2018;9(1):265.
Ning T, Zhang K, Heng BC, et al. Diverse effects of pulsed electrical stimulations on cells – with a focus on chondrocytes and cartilage regeneration. European Cells Mater. 2019;38:79–93.
Bai F, Gusbeth C, Frey W, et al. Nanosecond pulsed electric fields trigger cell differentiation in Chlamydomonas reinhardtii. Biochim Biophys Acta Biomembr. 2017;1859(5):651–61.
Morotomi-Yano K, Uemura Y, Katsuki S, et al. Activation of the JNK pathway by nanosecond pulsed electric fields. Biochem Biophys Res Commun. 2011;408(3):471–6.
Semenov I, Xiao S, Pakhomov AG. Primary pathways of intracellular Ca2+ mobilization by nanosecond pulsed electric field. Biochim Biophys Acta Biomembr. 2013;1828(3):981–9.
Rohmah MK, Lyrawati D, Yano K. Nanosecond pulsed electric fields (nsPEFs) induce Socs1 and Socs3 but not Socs2 gene expressions in Hela S3 cells. Jurnal Kedokteran Brawijaya. 2015;28(3):163–9.
Chopinet L, Rols M-P. Nanosecond electric pulses: a mini-review of the present state of the art. Bioelectrochemistry. 2015;103:2–6.
He L, Xiao D, Feng J, et al. Induction of apoptosis of liver cancer cells by nanosecond pulsed electric fields (nsPEFs). Med Oncol. 2017;34(2):24.
Wu S, Wang Y, Guo J, et al. Nanosecond pulsed electric fields as a novel drug free therapy for breast cancer: an in vivo study. Cancer Lett. 2013;343(2):268–74.
Zou H, ., Gan XL, Linghu LJ et al. Intense nanosecond pulsed electric fields promote cancer cell apoptosis through centrosome-dependent pathway involving reduced level of PLK1. Int J Gynecol Obstet 2012;119(2):S658-S658.
Zhang K, Guo J, Ge Z, et al. Nanosecond pulsed electric fields (nsPEFs) regulate phenotypes of chondrocytes through Wnt/β-catenin signaling pathway. Sci Rep. 2014;4:5836.
Ning T, Guo J, Zhang K, et al. Nanosecond pulsed electric fields enhanced chondrogenic potential of mesenchymal stem cells via JNK/CREB-STAT3 signaling pathway. Stem Cell Res Ther. 2019;10(1):1–12.
Baek S, Quan X, Kim S, et al. Electromagnetic fields mediate efficient cell reprogramming into a pluripotent state. ACS Nano. 2014;8(10):10125–38.
Wang J, Guo J, Wu S, et al. Synergistic effects of nanosecond pulsed electric fields combined with low concentration of gemcitabine on human oral squamous cell carcinoma in vitro. PLoS One. 2012;7(8):e43213.
Tsai C-C, Su P-F, Huang Y-F, et al. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol Cell. 2012;47(2):169–82.
Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. Int Soc Cell Ther Position State. 2006;8(4):315–7.
Yuin-Han L, Qiang W, Joon-Lin C, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–40.
Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392(10149):777–86.
Bhutani N, Brady JJ, Damian M, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463(7284):1042–7.
Ning T, Zhang K, Heng BC et al. Diverse effects of pulsed electrical stimulation on cells - with a focus on chondrocytes and cartilage regeneration. Eur Cells Mater. 2019;38:79–93.
Gianulis EC, Labib C, Saulis G, et al. Selective susceptibility to nanosecond pulsed electric field (nsPEF) across different human cell types. Cell Mol Life Sci. 2016;74(9):1–14.
Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–91.
Looijenga LHJ, Hans S, Leeuw HPJC, De et al. POU5F1 (OCT3/4) identifies cells with pluripotent potential in human germ cell tumors. Cancer Res. 2003;63(9):2244–50.
Ming LT, Nan WY, Min GX, et al. Effects of ectopic Nanog and Oct4 overexpression on mesenchymal stem cells. Stem Cells Dev. 2009;18(7):1013–22.
Hyslop L, Stojkovic M, Armstrong L, et al. Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells. 2005;23(8):1035–43.
Kusuyama J, Hwan SC, Ohnishi T, et al. 10. Low-intensity pulsed ultrasound (LIPUS) stimulation helps to maintain the differentiation potency of mesenchymal stem cells by induction in Nanog protein transcript levels and phosphorylation. J Orthop Trauma. 2016;30(8):S4–S5.
Ding H, Chen S, Yin JH, et al. Continuous hypoxia regulates the osteogenic potential of mesenchymal stem cells in a time-dependent manner. Mol Med Rep. 2014;10(4):2184–90.
Han S-M, Han S-H, Coh Y-R, et al. Enhanced proliferation and differentiation of Oct4- and Sox2-overexpressing human adipose tissue mesenchymal stem cells. Exp Mol Med. 2014;46(6):e101.
Marta G, Jochen U. From skin to the treatment of diseases--the possibilities of iPS cell research in dermatology. Exp Dermatol. 2011;20(6):523–8.
Cowan CA, Jocelyn A, Melton DA, et al. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309(5739):1369–73.
Freberg C, Dahl J, S, Collas P. Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract. Mol Biol Cell 2007;18(5):1543–1553.
Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol. 2004;6(10):984–90.
Zhao L, Yu Y, Deng C. Protein and mRNA expression of Shh, Smo and Gli1 and inhibition by cyclopamine in hepatocytes of rats with chronic fluorosis. Toxicol Lett. 2014;225(2):318–24.
Jing-Yu L, Min-Tie P, Ryutaro H, et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27(24):8748–59.
Liu Y, Mukhopadhyay P, Pisano MM, et al. Repression of Zeb1 and hypoxia cause sequential mesenchymal-to-epithelial transition and induction of aid, Oct4, and Dnmt1, leading to immortalization and multipotential reprogramming of fibroblasts in spheres. Stem Cells. 2013;31(7):1350–62.
Sen GL, Reuter JA, Webster DE, et al. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463(7280):563–7.
Mikkelsen TS, Jacob H, Xiaolan Z, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454(7200):49–55.
Danwei H, Kenji O, René M, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26(11):1269–75.
Shi Y, Desponts C, Do JT, et al. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3(5):568–74.
Zhu X, Chen F, Lu K et al. PPARgamma preservation via promoter demethylation alleviates osteoarthritis in mice. Ann Rheum Dis 2019;78(10):1420–9.
Acknowledgements
The authors would like to thank Mr. Kaile Wang for operating the nsPEFs equipment.
Funding
This work was supported by a National Natural Science Foundation of China grant (81772334).
Author information
Authors and Affiliations
Contributions
Kejia Li: conception and design, collection and assembly data, data analysis and interpretation, and manuscript writing. Tong Ning: conception and design, collection and assembly data, and data analysis and interpretation. Hao Wang: DNMT1 overexpression. Yangzi Jiang: data analysis and interpretation, and manuscript writing. Jue Zhang: conception and design, and final approval of manuscript. Zigang Ge: conception and design, data analysis and interpretation, and manuscript writing. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Animal experiments were approved by the Institutional Animal Care and Use Committee of Peking University, while human samples were harvested with approval of People’s Hospital of Peking University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1 :
Supplementary Methods and Materials.
Additional file 2 : Supplementary Tables
. Table S1. Primers for qRT-PCR. Table S2. Primers for BSP. Table S3. Primers for PCR.
Additional file 3 : Figure S1
. nsPEFs Enhance trilineage differentiation of MSCs. (A-C) qRT-PCR for the expressions of functional genes of trilineage differentiation for 14 days differentiation. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p≤0.05; **p≤0.01, ***p≤0.001, ****p≤0.0001, NS, p>0.05).
Additional file 4 : Figure S2
. nsPEFs have little effect on proliferation of MSCs. (A) Cell Viability based on MTT assay. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, NS, p>0.05) (B) Effect of nsPEFs on Cell cycle progression of MSCs. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA). (C) Colony-forming unit assay for MSCs stimulated by nsPEFs. (D) Viable colony counts. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 3 technical repeats, one-way ANOVA, NS, p>0.05).
Additional file 5 : Figure S3
. nsPEFs with varied parameters incur different effects on gene expressions of pluripotency genes of stem cells. (A-D) qRT-PCR for the expression of OCT4 and NANOG of pMSCs over 7 days after stimulation by nsPEFs. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p≤0.05; **p≤0.01, ***p≤0.001, ****p≤0.0001, NS, p>0.05) (E and F) qRT-PCR for the expressions of OCT4 and NANOG of ESCs at 2 hours after stimulation by nsPEFs. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, *p≤0.05; **p≤0.01, ***p≤0.001, ****p≤0.0001, NS, p>0.05).
Additional file 6 : Figure S4
. nsPEFs have no significant effect on DNMT3a/b. (A) Western blot for DNMT3a/b expression level of pMSCs at 2 hours after stimulation by nsPEFs. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, NS, p>0.05). (B) Western blot for DNMT3a/b expression level of hMSCs at 2 hours after stimulation by nsPEFs. (3 batches of studies were tested with 3 biological donors, values are mean ± SEM from one representative batch with 5 technical repeats, one-way ANOVA, NS, p>0.05).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, K., Ning, T., Wang, H. et al. Nanosecond pulsed electric fields enhance mesenchymal stem cells differentiation via DNMT1-regulated OCT4/NANOG gene expression. Stem Cell Res Ther 11, 308 (2020). https://doi.org/10.1186/s13287-020-01821-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13287-020-01821-5