Abstract
Background
Tuberous sclerosis complex (TSC) is a rare monogenic disorder characterized by benign tumors in multiple organs as well as a high prevalence of epilepsy, intellectual disability and autism. TSC is caused by inactivating mutations in the TSC1 or TSC2 genes. Heterozygocity induces hyperactivation of mTOR which can be inhibited by mTOR inhibitors, such as rapamycin, which have proven efficacy in the treatment of TSC-associated symptoms. The aim of the present study was (1) to identify molecular changes associated with social and cognitive deficits in the brain tissue of Tsc1 +/− mice and (2) to investigate the molecular effects of rapamycin treatment, which has been shown to ameliorate genotype-related behavioural deficits.
Methods
Molecular alterations in the frontal cortex and hippocampus of Tsc1 +/− and control mice, with or without rapamycin treatment, were investigated. A quantitative mass spectrometry-based shotgun proteomic approach (LC-MSE) was employed as an unbiased method to detect changes in protein levels. Changes identified in the initial profiling stage were validated using selected reaction monitoring (SRM). Protein Set Enrichment Analysis was employed to identify dysregulated pathways.
Results
LC-MSE analysis of Tsc1 +/− mice and controls (n = 30) identified 51 proteins changed in frontal cortex and 108 in the hippocampus. Bioinformatic analysis combined with targeted proteomic validation revealed several dysregulated molecular pathways. Using targeted assays, proteomic alterations in the hippocampus validated the pathways “myelination”, “dendrite,” and “oxidative stress”, an upregulation of ribosomal proteins and the mTOR kinase. LC-MSE analysis was also employed on Tsc1 +/− and wildtype mice (n = 34) treated with rapamycin or vehicle. Rapamycin treatment exerted a stronger proteomic effect in Tsc1 +/− mice with significant changes (mainly decreased expression) in 231 and 106 proteins, respectively. The cellular pathways “oxidative stress” and “apoptosis” were found to be affected in Tsc1 +/− mice and the cellular compartments “myelin sheet” and “neurofilaments” were affected by rapamycin treatment. Thirty-three proteins which were altered in Tsc1 +/− mice were normalized following rapamycin treatment, amongst them oxidative stress related proteins, myelin-specific and ribosomal proteins.
Conclusions
Molecular changes in the Tsc1 +/− mouse brain were more prominent in the hippocampus compared to the frontal cortex. Pathways linked to myelination and oxidative stress response were prominently affected and, at least in part, normalized following rapamycin treatment. The results could aid in the identification of novel drug targets for the treatment of cognitive, social and psychiatric symptoms in autism spectrum disorders. Similar pathways have also been implicated in other psychiatric and neurodegenerative disorders and could imply similar disease processes. Thus, the potential efficacy of mTOR inhibitors warrants further investigation not only for autism spectrum disorders but also for other neuropsychiatric and neurodegenerative diseases.
Similar content being viewed by others
Background
Tuberous sclerosis complex (TSC) is a rare multi-system monogenic hamartomatous disorder, which is caused by mutations inactivating the TSC1 (hamartin) or TSC2 (tuberin) genes. TSC is characterized by benign tumors in multiple organs, including the brain, kidneys, heart and eyes [1]. Over 90% of TSC patients develop epilepsy, and around 50% present with neuropsychiatric problems, such as intellectual disability (50%) [2, 3], autism spectrum disorder (ASD) (17–68%), schizophrenia (10–30%) and anxiety disorders (40%) [4], which account for most of the mortality and morbidity [5].
At the molecular level, both Tsc1 and Tsc2 protein products form hetero-dimers which inhibit the GTP-binding protein RHEB (Ras homolog enriched in the brain). Consequently, mutations within either Tsc1 or Tsc2 lead to increased levels of activated RHEB [6], which causes hyperactivation of mammalian target of rapamycin (mTOR) signaling, a constitutive phosphorylation of eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) and activation of ribosomal protein S6 through S6K1 phosphorylation [7, 8]. The net effect is enhanced protein translation, cell proliferation and growth [9]. Notably, increased mTOR signaling and subsequent changes in global protein synthesis are shared molecular mechanisms of several rare neurodevelopmental disorders with an increased prevalence of ASD, such as fragile X syndrome (FXS) [10].
The hyperactivation of mTOR induced by Tsc1 and Tsc2 heterozygosity can be inhibited by mTOR inhibitors, such as the macrolide rapamycin. Rapamycin is an immunosuppressant, which is widely prescribed to prevent rejection in organ transplantation and exerts anti-tumor properties [11–13]. Rapamycin binds FK-binding protein 12 (FKBP12), and as a complex, rapamycin-FKBP12 directly binds to the mTOR complex 1 (mTORC1), thus reducing phosphorylation of downstream mTOR targets [14, 15]. Rapamycin and other mTOR inhibitors have been shown to be efficacious in the treatment of several TSC-associated tumors as well as seizures [16–19] and may ameliorate the symptoms of neurodevelopmental disorders in adults [20, 21]. In TSC mouse models, rapamycin limits tumor growth [22, 23], reduces neuropathology and ameliorates epileptic seizures as well as learning deficits [24–26]. It was recently reported that rapamycin normalizes social interaction deficits relevant to core disabilities associated with ASD in both Tsc1 +/− and Tsc2 +/− mice [27].
Here, we investigated the Tsc1 +/− mouse model, which exhibits haploinsufficiency for the Tsc1 gene, in an attempt to identify molecular changes associated with the neuropsychiatric phenotype of TSC patients [5]. In this mouse model, the typical human cerebral pathology of spontaneous seizures, cerebral lesions and giant dysmorphic cells could not been detected using immuno-cytochemistry and high resolution magnetic resonance imaging, respectively [28]. Furthermore, spine number and dendritic branching are normal [28]. However, the Tsc1 +/− mouse shows prominent behavioural deficits which mimic core symptoms of ASD and other neuropsychiatric disorders [28]. Tsc1 +/− mice show hippocampal learning deficits using the Morris water maze test and contextual fear conditioning, as well as social deficits indicated by reduced social interaction and nest building [28]. Consequently, the Tsc1 +/− mouse is a suitable model to investigate aspects of the molecular pathology associated with neuropsychiatric spectrum disorders, especially in relation to ASD and intellectual disability. In this study, we attempted to identify changes in molecular pathways in the frontal cortex and hippocampus of the Tsc1 +/− mouse model using a mass spectrometry-based proteomics approach. We also investigated protein changes associated with rapamycin treatment. Findings from this study could aid in the identification of novel drug targets for the treatment of cognitive, social and psychiatric symptoms in ASD.
Methods
A more detailed description of the materials and methods used in this study can be found in the supplementary methods section (Additional file 1).
Animals
Tsc1 +/− mice were generated by replacing exons 6 through to 8 of the Tsc1 gene with a selection cassette, as described previously [29]. This leads to the generation of Tsc1 null embryos which express Tsc1 transcripts in which exon 5 and 9 are fused, leading to a premature TGA stop codon. Consequently, any protein translated from this allele lacks all of the known functional domains of hamartin including the putative Rho activation domain. The Tsc1 +/− mutant mouse was crossed six times into the C57BL/6J OlaHsD background and then at least three times into the C57BL/6N/HsD background. The offspring consisted of Tsc1 +/− mice and wildtype littermates. Mice were genotyped when they were about 7 days old. They were housed in groups and kept on a 12-h light/dark cycle, with food and water available ad libitum. Mice were culled when they were 6–8 weeks old and genotype groups were sex- and age-matched for the experiments for consistency with the published behavioural data [28]. Mouse genotypes were blinded using codes all the way through to the sample preparation stage. The codes were un-blinded for the mass spectrometry analysis since samples had to be distributed evenly to avoid run time biases. All animal experiments were approved by the Dutch Ethical Committee or in accordance with Institutional Animal Care and Use Committee guidelines.
Rapamycin treatment
Mice were injected intraperitoneally with 5 mg/kg rapamycin or vehicle for 5 days and culled 24 h after the last injection [27]. Rapamycin was dissolved in 5% dimethyl sulfoxide diluted with saline to 5 ml/kg. Mice were 5–7 weeks old at the time of injection.
Proteomic sample preparation
Sample preparation was carried out as described previously [30–32]. Based on the lysates, two randomized, blinded, independent sample preparations were prepared for liquid chromatography mass spectrometry (in expression mode; LC-MSE) and selected reaction monitoring mass spectrometry to avoid bias in sample preparations.
Label-free LC-MSE proteomic profiling of brain tissue
Brain tissue analysis and data processing were performed as described previously [31, 33, 34]. The Swiss-Prot human reference proteome (Uniprot release March 2013, 20,252 entries) was used for protein identification searches. Protein abundance changes for the comparisons between Tsc1 +/− and wildtype were determined by the MSstats package [35] based on mixed-effect models using the peptide intensities, following log2 transformation and exclusion of intensity values deviating by more than 3 standard deviations from the mean of each group.
Protein set enrichment analysis
Significantly changed proteins were partitioned into three bins, according to their ratio: proteins decreased in abundance (ratio < 1.0), proteins increased in abundance (ratio > 1.0) and a bin to identify general disturbed pathways which included all proteins with increased and decreased abundance (ratio > 1 and <1). The R package database org.mouse.eg.db version 2.8.0 was used for gene ontology (GO) term annotation based on entrez gene identifiers and GO-term enrichment analysis was performed using GOstats.
Label-based SRM mass spectrometry
Abundance alterations of a panel of 43 candidate proteins previously implicated in the Tsc1 +/− mouse pathology were measured using a targeted SRM mass spectrometry approach as described previously [32, 36] following the guidelines of Lange et al. [37]. SRMstats was used at default settings [37]. The final transitions, collision energy and retention time windows used for each peptide can be requested.
Results
Label-free LC-MSE proteomic profiling of Tsc1 +/− mouse brains
We investigated protein abundance changes in the frontal cortex and hippocampus of the Tsc1 +/− mouse. LC-MSE analysis resulted in the identification of 522 proteins (7071 peptides) in the frontal cortex and 463 proteins (5149 peptides) in the hippocampus. Of these, the levels of 51 proteins were altered in the frontal cortex (FDR-adjusted *p < 0.05) and 108 in the hippocampus (FDR-adjusted *p < 0.05). In the frontal cortex, 17 of the changed proteins were altered by more than 10%, as were 49 of the 108 changed hippocampal proteins (Additional file 2). In the case of the frontal cortex, this included adenylyl cyclase-associated protein 2 (CAP2, ratio = 0.89, FDR-adjusted *p = 0.013), elongation factor 1–α2 (EIF1A2, ratio = 0.97, FDR-adjusted *p = 0.03), eukaryotic translation initiation factor 3 subunit L (eIF3l, ratio = 1.17, FDR-adjusted *p = 0.03) and elongation factor 2 (Eef2, ratio = 0.95, FDR-adjusted *p = 0.05), which are all regulators of translation. Copine 6 (ratio = 1.1, FDR-adjusted p = 0.0097) and copine 8 (ratio = 0.8, FDR-adjusted p = 3.9 × 10−6), which are associated with synaptic plasticity, were changed in the hippocampus. Nine proteins (NCDN⬇⬇, MAP2⬆⬆, SUCB1⬇⬇, MYPR⬆⬇, NDUS7⬇⬇, DPYL2⬇⬇, AT1A2⬇⬆, CRYM⬇⬆, ARP3⬆⬇) were found to be changed in both frontal cortex (first arrow) and hippocampal tissue (second arrow) (Additional file 2).
Gene set enrichment analysis was employed to investigate if the altered 108 and 51 proteins were enriched in biological pathways and cellular compartments. Based on GO enrichment analysis, proteins responsible for the biological pathways “reproductive behaviour” (p = 0.008), “neurological system process” (p = 0.010) and “visual learning” (p = 0.028) were altered in the frontal cortex of the Tsc1 +/− mouse. In the hippocampus, the proteins were related to the biological pathways “ribonucleotide energy metabolism” (p = 0.0097), “protein polymerisation” (p = 0.005) and “oxidative stress” (p = 0.009). One pathway, “visual learning”, was identified in both the frontal cortex and hippocampus proteomic analyses. Cellular compartment GO association enrichment revealed that the altered proteins were associated with “myelination” and “dendrite” in the frontal cortex and “myelin sheet” and “endoplasmic reticulum-Golgi intermediate compartment” in the hippocampus.
Selected reaction monitoring (SRM) validation of Tsc1 +/− brain proteomic alterations
For orthogonal proteomic validation of the proteomics results, we employed a targeted label-based LC-SRM approach to specifically quantify the levels of 43 candidate proteins derived from LC-MSE profiling, subsequent pathway analysis, literature findings and already established in-house assays. Label-based SRM assays are highly specific and sensitive and outperform immunoblotting analyses for validation [38–41]. This analysis showed that the levels of 5 and 20 of the targeted proteins were significantly changed in the frontal cortex and hippocampus, respectively (p < 0.05, Table 1). Specifically, two proteins (MYPR, PURA) out of five LC-MSE candidates were validated in the frontal cortex and six (NSF, MYPR, MBP, CPNE6, SODC, MARCS) out of ten LC-MSE-derived protein candidates were validated in the hippocampus (Table 1). Using targeted SRM-assays, we confirmed the more prominent proteomic alterations in the hippocampus and further validated GO-terms “myelination”, “dendrite” and “oxidative stress” through confirmation of changes in MYPR, MBP, TSN2 (all myelin specific proteins), MAP2 (dendritic marker) and SODC (oxidative stress marker). We further detected an upregulation in ribosomal proteins and mTOR kinase in the Tsc1 +/− mice (Table 1).
Label-free LC-MSE proteomic profiling of the Tsc1 +/− hippocampus following rapamycin treatment
To investigate the effects of rapamycin on the brain proteome, label-free LC-MSE analysis was employed on Tsc1 +/− and wildtype mice treated with rapamycin or vehicle (Fig. 1a). Only the hippocampus was studied in this case as this brain region was more affected with regard to significantly changed proteins (Table 1). The hippocampus plays not only an important role in cognition, but hippocampal dysfunction has also been linked to a wide range of neuropsychiatric symptoms [42, 43]. Deficits in consolidating short- and long-term memory and spatial navigation have been shown to be impaired in Tsc1 +/− and Tsc2+/− mice and were reversed by rapamycin treatment in Tsc2+/− mice [24]. LC-MSE analysis led to the identification of 8648 total peptides which translated to 597 proteins, which were detected across all samples. Interestingly, rapamycin treatment exerted a stronger proteomic effect in Tsc1 +/− compared to wildtype mice (Fig. 1c (2 and 4)) with significant changes in 231 and 106 proteins, respectively. An overall decrease in protein levels was found in both Tsc1 +/− and wildtype mice.
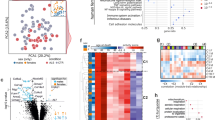
Label-free LC-MSE and Label-based SRM analysis of Tsc1+/− and wildtype mice under rapamycin and vehicle treatment identifies distinct proteomic changes. a) Flow chart of the experimental design. b) Significant protein changes in the hippocampus of rapamycin-treated WT and Tsc1 +/− mice identified by label-free LC-MSE. Orange dots refer to proteins that are increased in abundance, and green dots represent downregulated proteins following rapamycin treatment. c 2 and 4 (see below) refer to significantly changed proteins following rapamycin treatment in wildtype (“treatment effect in Wt mice”) or Tsc1+/− mice (“treatment effect in Tsc1 +/− mice”). Protein enrichment analysis was performed on the identified proteins. Yellow and blue dots represent the TSC genotype effect following rapamycin or vehicle treatments (b 1 and 2) and are linked to c 1 and 3. Proteins changing due to a combination of TSC genotype and rapamycin treatment are labeled black or purple, respectively. b 1 Rapamycin induced changes in wildtype mice as compared to Tsc1 +/− mice following vehicle treatment. b 2 Rapamycin induced changes in Tsc1 +/− mice compared to Tsc1 +/− alterations following rapamycin treatment. c) Bar plots of genotype and treatment effects identified through global protein profiling and significantly changed proteins identified in the targeted SRM analysis. Number of significant proteins, percentage of up- and downregulated proteins and enriched pathways linked to the up- and downregulated proteins are displayed
Next, proteins were tested which were affected in all four comparisons. This showed that 9 proteins were changed in common (FRM4A, PEA15, PERQ1, MAP2, BASP, CLD11, ALBU, TCAL3, CLH) and that the levels of 54 proteins were affected by rapamycin treatment in both wildtype and Tsc1 +/− mice; of these proteins, 52 corresponded in their fold change direction (37 of the 52 proteins were decreased in abundance and 15 increased, respectively). Pathway analysis linked the 52 overlapping proteins to the biological process of “translation” (p = 0.00082), “macromolecule biosynthetic process” (p = 0.005) and “gene expression” (p = 0.014). Using KEGG (Kyoto Encyclopedia of Genes and Genomes) annotation, “ribosome” was the most significant pathway (p = 1.7 × 10−7) in the enrichment analysis.
We further employed enrichment analysis for the genotype comparisons and the treatment comparisons (Fig. 1c (1–4)). This associated the biological pathways “oxidative stress” and “apoptosis” with the significantly changed proteins identified in the Tsc1 +/− vs Wt comparison (Fig. 1c (1)). Furthermore, cellular compartments of myelin sheet and neurofilaments were affected by rapamycin treatment in both Tsc1 +/− and Wt mice (Fig. 1c (2 and 4)). Proteins with decreased levels due to rapamycin treatment mostly related to the biological pathways “translation”, “macromolecular complex assembly” and “chromosome organization” (Fig. 1c (2 and 4)). Downregulation of the pathway “chromosome organization” was specifically observed in Tsc1 +/− mice following rapamycin treatment (Fig. 1c (4)).
Importantly, 41 proteins which were altered in vehicle-treated Tsc1 +/− mice were normalized following rapamycin treatment. These proteins include a set of proteins where rapamycin treatment normalizes the genotype-induced protein alterations to wildtype levels (33 proteins) and a set of proteins where rapamycin normalizes the genotype-effect below or above baseline levels (8 proteins). The former include the Glycine receptor subunit alpha-4 (GLRA4), the Calcium-dependent secretion activator 1 (CAPS1), Rod cGMP-specific 3′,5′ cyclic phosphodiesterase beta (PDE6B) and Guanine deaminase (GUAD) (Fig. 2 (N)); the latter include Rho-associated protein kinase 2 (ROCK2) and ribosomal proteins (RS18, RL4, RS9). All ribosomal proteins affected by rapamycin treatment were decreased in their abundance levels. Furthermore, proteins were identified that are affected by rapamycin treatment in both wildtype and mutant mice, although there was no difference in their abundance levels between vehicle-treated mutant and wildtype mice (Fig. 2 (R)). This set was comprised of 41 proteins. Amongst them are the anaphase promoting complex s7 (APC7), calcineurin subunit B type 1 (CANB1) and the GABA aminotransferase (GABT). Finally, the levels of six proteins were found to be altered between mutant and wildtype but did not change following rapamycin treatment. Neuromodulin (NEUM), the excitatory amino acid transporter 2 (EAAT2) and SMP25 are examples.
Multivariate analysis of LC-MSE estimates as shown by condition plots illustrating the differences between the WT and Tsc1 +/− mice with and without rapamycin treatment. X-axis is condition and y-axis is log ratio of endogenous (L light) over reference (H heavy) peptides. Dots represent the mean of the log2 ratio for each condition, and error bars indicate the confidence intervals with 0.95 significance. The interval is not related to the model based analysis. Significant changes as measured by LC-MSE are indicated below each protein. Illustrated are examples of proteins which are affected by rapamycin treatment and which are normalized by rapamycin treatment. Corrected p values (p*) were determined by post hoc correction after Benjamini-Hochberg [91]. CR = Wt + rapamycin, CS = Wt + vehicle, TR = Tsc1 +/− + rapamycin, TS = Tsc1 +/− − rapamycin
SRM validation of rapamycin treatment effects in the Tsc1 +/− hippocampus
The next phase of the study involved a targeted proteomic approach to validate the findings of the rapamycin study (see Fig. 1c (1–4) [targeted]). This focused on myelination deficits, alterations in the translational machinery and proteins found to be altered in the label-free LC-MSE discovery study of the Tsc1+/− mouse (SODC, NSF, MAP2, PKCG). The rapamycin-target mTOR was included as a positive control.
This analysis resulted in validation of increased myelin-associated proteins in Tsc1 +/− compared to wildtype mice (Fig. 1c (1 and 3)). Furthermore, the decrease in oxidative stress-related proteins in the Tsc1 +/− mouse was validated by confirming decreased levels of SODC (Fig. 1c (1)). Additionally, altered levels of NSF and PKCG were also validated (Fig. 1c [targeted] and Fig. 3).
Multivariate analysis of SRM estimates as shown by condition plots illustrating the differences between Wt and Tsc1 +/− mice with and without rapamycin treatment. X-axis represents the condition and the y-axis the log ratio of endogenous (L light) over reference (H heavy) peptides. Dots represent the mean of log2 ratio for each condition, and error bars indicate the confidence intervals with 0.95 significance. The interval is not related to the model based analysis. Significant changes as measured by LC-MSE are indicated below each protein. SRM was able to validate protein abundance changes identified by label-free LC-MSE. Corrected p values (p*) were determined by post hoc correction after Benjamini-Hochberg [91]
In the case of rapamycin treatment effects, decreased levels of proteins involved in translation could be validated. Specifically, decreased levels of all three tested ribosomal subunits as well as the mTOR-kinase were identified. Moreover, the analysis showed that rapamycin normalized the protein levels of SODC, NSF and PKCG in Tsc1 +/− mice (Fig. 1c [targeted]).
Discussion
The pathogenesis of psychiatric disorders such as ASD remains elusive, and there is accumulating evidence that several neuronal circuits and pathways are affected. This is especially true for the social, cognitive and neuropsychiatric symptoms associated with these disorders. In an attempt to gain further insight into these pathways, this study combines unbiased and targeted proteomic approaches to investigate the hippocampus and frontal cortex of a mouse model of TSC, which is one of the most frequent causes of syndromic ASD [44]. The investigated Tsc1 +/− mouse model exhibits social and cognitive deficits, which are core behavioural symptoms of ASD in humans [45] and other relevant rodent models [46], without any obvious brain pathology (such as tumors or epilepsy). This makes the Tsc1 +/− mouse an excellent model of pharmacologically treatable ASD. A previous study has demonstrated the effectiveness of rapamycin to normalize reciprocal social interactions in this model [27]. The aim of the present study was to investigate proteins and pathways affected by rapamycin treatment, which could support drug discovery efforts and in turn the development of improved treatments for TSC, ASD and possibly other neuropsychiatric disorders.
Proteomic profiling of the frontal cortex and hippocampus brain tissue in this study identified and quantified a large number of significantly changed proteins in the Tsc1 +/− mouse model. Differentially expressed proteins and altered molecular pathways were identified and selected candidate proteins were validated using SRM as a highly quantitative method. In a second stage, proteomic analysis of rapamycin treatment effects were investigated to identify down-stream effects of mTOR-pathway inhibition in the hope to gain new insights into the molecular underpinnings of social impairments in ASD and other psychiatric disorders.
We were able to show that myelin proteins and the translational machinery, specifically several ribosomal subunits, were significantly altered in Tsc1 +/− mice treated with rapamycin. Our findings of lower ribosomal subunit abundances are consistent with a rapamycin-induced downregulation of ribosomal biogenesis [47]. Regarding the effects on myelination, previous research has linked the mTOR pathway to oligodendrocyte differentiation and axonogenesis [48]. Oligodendrocytes produce myelin, and this is specifically regulated at the late progenitor to immature oligodendrocyte transition stage (as shown by changes in expression of the myelin marker proteins MYPR and MBP). We identified an increase in myelin proteins in the Tsc1 +/− mouse, but not in Wt mice. A recent study has shown that ablation of TSC1 is associated with oligodendrocyte-specific over-activation and subsequent hypomyelination [49]. An increase in myelin proteins in the Tsc1 +/− mouse brain may be due to the globally enhanced (Table 1) protein translation and cell proliferation in the context of mTOR hyperactivation. An increase in cell growth and proliferation could interfere with oligodendrocyte maturation and thus result in incomplete myelination as seen in some demyelinating diseases, such as multiple sclerosis [50].
MYPR and MBP as well as TSN-2 were amongst the altered myelin proteins. These proteins play an important role in oligodendrocyte differentiation during development. Furthermore, in vitro studies have shown that MBP mRNA and protein expression are significantly decreased by mTOR inhibition [48, 51]. Inhibiting mTOR in oligodendrocyte precursor cell/dorsal root ganglion co-cultures potently abrogated oligodendrocyte differentiation and reduced numbers of myelin segments. Disorganized and structurally compromised axons with poor myelination have already been found in TSC patients, and this may at least to some extent explain the behavioural and cognitive deficits associated with the disorder [52]. Impaired adult myelination has been shown in the prefrontal cortex of socially isolated mice [53]. Importantly, changes in oligodendrocyte function and myelination abnormalities are amongst the most consistent hallmarks of psychiatric pathology in post-mortem brain studies. Changes were reported for schizophrenia, bipolar disorder, depression and ASD [36, 54–58]. In wildtype mice, rapamycin treatment led to a reduction of myelin and myelin protein expression [59, 60]. This is consistent with our findings, where rapamycin affects both wildtype (reduced) and mutant myelin (increased) protein expression.
Interestingly, several proteins that we found altered in the Tsc1 +/− mouse model were reversed by rapamycin treatment. One of these proteins, a glycine receptor subunit, which abundance was decreased in the mutant and normalized by rapamycin treatment, could be a potential drug target for novel treatments of ASD and schizophrenia-spectrum disorders. The glycine receptor co-localizes with GABAA receptors on hippocampal neurons [61]. A microdeletion at Xq22.2 implicates GLRA4 to be involved in intellectual disability and behavioural problems [62]. In a case report, glycine receptor antibodies could be detected in a patient with treatment-resistant focal epilepsy, tantrums, clumsiness and impaired speech [63] and in patients with progressive encephalomyelitis with rigidity and myoclonus stiff person syndrome [64]. Treatments targeting the glycine transporter are under investigation as novel treatment approaches for schizophrenia [65].
Other altered proteins, which are normalized by rapamycin treatment, included the calcium-dependent secretion activator 1 (CAPS1) (decreased in Tsc1 +/− and normalized by rapamycin), two guanine metabolism associated proteins (guanine deaminase and PDE6B; both increased in Tsc1 +/− and normalized by rapamycin) and the vesicle-fusing ATPase NSF (increased in Tsc1 +/−, normalized by rapamycin), a molecular component of the exocytosis machinery [66], which is required for membrane fusion [67] and regulates the disassembly of SNARE complexes on early endosomes [68]. The NSF gene has also been linked with cocaine dependence [69] and schizophrenia [36, 70]. Direct interactions with cell surface receptors such as AMPA receptors [71, 72], β2-adrenergic receptors [73], dopaminergic receptors [74] and the adrenomedulin receptor [75] have been reported. Interestingly, a coordinated action of NSF and PKC regulates GABAB receptor signaling efficiency [76]. PKCG was found to be strongly downregulated by rapamycin treatment in this study and is known to be involved in the regulation of the neuronal receptors GLUR4 and NMDAR1 [77]. It binds and phosphorylates the GLUR4 glutamate receptor and regulates its function by increasing membrane-associated GRIA4 expression [78]. Several preclinical and clinical trials have investigated mGLUR antagonists for the treatment of social deficits in ASD [78, 79] and ASD associated with FXS [80, 81] and PKCG inhibitors could represent a novel treatment strategy to ameliorate cognitive and social deficits. Notably, Ketamine, which is thought to exert antidepressant action through modulation of mTOR pathway activity [82], potentiates persistent learning and memory impairment through the PKCG-ERK signaling pathway [83].
Another protein strongly downregulated following rapamycin treatment is the anaphase promoting complex S7 (APC7), which is a cell cycle-regulated E3 ubiquitin ligase controlling progression through mitosis and the G1 phase of the cell cycle. The control of APC7 through rapamycin might be a major breakpoint in cell proliferation. Rapamycin has already been shown to also downregulate the expression of the APC/C inhibitor Emi1 [84].
Copine 6, which we found upregulated in the hippocampus of Tsc1 +/− compared to wildtype mice, is a calcium-dependent regulator of the actin cytoskeleton in neuronal spines and negatively regulates spine maturation during neuronal development [85]. Changes in copine 6 expression may be involved in neurodevelopmental disorders, as deformed dendritic spines and changes in spine density are hallmarks of many neurodevelopmental conditions, such as Down’s syndrome [86, 87] and FXS [88]. Interestingly, hippocampi from patients suffering from uncontrolled epileptic seizures, typically a problem in tuberous sclerosis patients, exhibit a decrease in spine density [89]. We also found that MAP2, a dendritic spine marker, was increased by Tsc1 heterozygocity and decreased by rapamycin treatment.
Interestingly, over twice as many significantly changed proteins were identified in the Tsc1 +/− hippocampus as compared to control animals following rapamycin treatment (Fig. 1c; 231 vs. 106 changed proteins, respectively). TSC1 mutations are linked to numerous changes in biochemical processes, including cell cycle regulation, translational control and metabolism which are linked to mTOR pathway hyperactivation. It can be speculated that rapamycin-related inhibition of the mTORC1 complex results in TSC genotype-dependent adaptations in a wide range of molecular pathways. These adaptations could be indirectly involved in the therapeutic effect of rapamycin. A further explanation for the enhanced rapamycin treatment effect in the Tsc1+−/ mice is selective vulnerability. Mutant mice might be more susceptible to the treatment as mTOR hyperactivation modulated similar downstream molecular pathways during neurodevelopment as are affected by the rapamycin-induced mTOR hypoactivation.
Conclusions
Taken together, the results from this comprehensive study represent the first proteomic characterization of the Tsc1 +/− mouse model to date. The findings yield novel insights into the molecular Tsc1 +/− mouse pathology as well as the molecular effects of rapamycin treatment, which is an effective treatment for several clinical symptoms of the tuberous sclerosis complex. Furthermore, the mTOR pathway, which is modulated by rapamycin treatment, is a novel drug target for the treatment of ASD, schizophrenia and affective disorders. We hope that the findings from this study will provide evidence and support for future clinical trials in the field of neuropsychiatric disorders.
Abbreviations
- ASD:
-
Autism spectrum disorder
- LC:
-
Liquid chromatography
- MBP:
-
Myelin basic protein
- mTOR:
-
Mammalian target of rapamycin
- MYPR:
-
Myelin proteolipid protein
- NSF:
-
Vesicle-fusing ATPase
- PKCG:
-
Protein kinase C gamma
- SODC:
-
Superoxide dismutase C
- SRM:
-
Selected reaction monitoring
- TSC:
-
Tuberous sclerosis complex
- TSC1:
-
Harmartin
- TSC2:
-
Tuberin
- TSN-2:
-
Tetraspanin 2
- Wt:
-
Wildtype
References
Islam MP, Roach ES. Tuberous sclerosis complex. Handb Clin Neurol. 2015;132:97–109.
de Vries PJ, Prather PA. The tuberous sclerosis complex. N Engl J Med. 2007;356(1):92. author reply 93-94
Joinson C, O'Callaghan FJ, Osborne JP, Martyn C, Harris T, Bolton PF. Learning disability and epilepsy in an epidemiological sample of individuals with tuberous sclerosis complex. Psychol Med. 2003;33(2):335–44.
Roach ES, Smith M, Huttenlocher P, Bhat M, Alcorn D, Hawley L. Diagnostic criteria: tuberous sclerosis complex. Report of the Diagnostic Criteria Committee of the National Tuberous Sclerosis Association. J Child Neurol. 1992;7(2):221–4.
Curatolo P, Moavero R, de Vries PJ. Neurological and neuropsychiatric aspects of tuberous sclerosis complex. Lancet Neurol. 2015;14(7):733–45.
Kwiatkowski DJ, Manning BD. Tuberous sclerosis: a GAP at the crossroads of multiple signaling pathways. Hum Mol Genet. 2005;14 Spec No. 2:R251–8.
Ehninger D, de Vries PJ, Silva AJ. From mTOR to cognition: molecular and cellular mechanisms of cognitive impairments in tuberous sclerosis. J Intellect Disabil Res. 2009;53(10):838–51.
Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5(12):931–42.
Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–45.
Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480(7375):63–8.
Samuels JA. Treatment of renal angiomyolipoma and other hamartomas in patients with tuberous sclerosis complex. Clin J Am Soc Nephrol. 2017;12(7):1196–1202.
Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009.
Phan AT, Dave B. The pivotal role of mammalian target of rapamycin inhibition in the treatment of patients with neuroendocrine tumors. Cancer Med. 2016;5(10):2953–64.
Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369(6483):756–8.
Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci U S A. 1995;92(11):4947–51.
Franz DN, Leonard J, Tudor C, Chuck G, Care M, Sethuraman G, Dinopoulos A, Thomas G, Crone KR. Rapamycin causes regression of astrocytomas in tuberous sclerosis complex. Ann Neurol. 2006;59(3):490–8.
Bissler JJ, McCormack FX, Young LR, Elwing JM, Chuck G, Leonard JM, Schmithorst VJ, Laor T, Brody AS, Bean J, et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med. 2008;358(2):140–51.
Davies DM, Johnson SR, Tattersfield AE, Kingswood JC, Cox JA, McCartney DL, Doyle T, Elmslie F, Saggar A, de Vries PJ, et al. Sirolimus therapy in tuberous sclerosis or sporadic lymphangioleiomyomatosis. N Engl J Med. 2008;358(2):200–3.
French JA, Lawson JA, Yapici Z, Ikeda H, Polster T, Nabbout R, Curatolo P, de Vries PJ, Dlugos DJ, Berkowitz N, et al. Adjunctive everolimus therapy for treatment-resistant focal-onset seizures associated with tuberous sclerosis (EXIST-3): a phase 3, randomised, double-blind, placebo-controlled study. Lancet. 2016;388(10056):2153–63.
Ehninger D, Li W, Fox K, Stryker MP, Silva AJ. Reversing neurodevelopmental disorders in adults. Neuron. 2008;60(6):950–60.
Citraro R, Leo A, Constanti A, Russo E, De Sarro G. mTOR pathway inhibition as a new therapeutic strategy in epilepsy and epileptogenesis. Pharmacol Res. 2016;107:333–43.
Kenerson HL, Aicher LD, True LD, Yeung RS. Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors. Cancer Res. 2002;62(20):5645–50.
Kenerson H, Dundon TA, Yeung RS. Effects of rapamycin in the Eker rat model of tuberous sclerosis complex. Pediatr Res. 2005;57(1):67–75.
Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, Kwiatkowski DJ, Ramesh V, Silva AJ. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med. 2008;14(8):843–8.
Zeng LH, Xu L, Gutmann DH, Wong M. Rapamycin prevents epilepsy in a mouse model of tuberous sclerosis complex. Ann Neurol. 2008;63(4):444–53.
Meikle L, Pollizzi K, Egnor A, Kramvis I, Lane H, Sahin M, Kwiatkowski DJ. Response of a neuronal model of tuberous sclerosis to mammalian target of rapamycin (mTOR) inhibitors: effects on mTORC1 and Akt signaling lead to improved survival and function. J Neurosci. 2008;28(21):5422–32.
Sato A, Kasai S, Kobayashi T, Takamatsu Y, Hino O, Ikeda K, Mizuguchi M. Rapamycin reverses impaired social interaction in mouse models of tuberous sclerosis complex. Nat Commun. 2012;3:1292.
Goorden SM, van Woerden GM, van der Weerd L, Cheadle JP, Elgersma Y. Cognitive deficits in Tsc1+/− mice in the absence of cerebral lesions and seizures. Ann Neurol. 2007;62(6):648–55.
Wilson C, Idziaszczyk S, Parry L, Guy C, Griffiths DF, Lazda E, Bayne RA, Smith AJ, Sampson JR, Cheadle JP. A mouse model of tuberous sclerosis 1 showing background specific early post-natal mortality and metastatic renal cell carcinoma. Hum Mol Genet. 2005;14(13):1839–50.
Wesseling H, Guest PC, Lee CM, Wong EH, Rahmoune H, Bahn S. Integrative proteomic analysis of the NMDA NR1 knockdown mouse model reveals effects on central and peripheral pathways associated with schizophrenia and autism spectrum disorders. Mol Autism. 2014;5:38.
Wesseling H, Want EJ, Guest PC, Rahmoune H, Holmes E, Bahn S. Hippocampal proteomic and metabonomic abnormalities in neurotransmission, oxidative stress, and apoptotic pathways in a chronic phencyclidine rat model. J Proteome Res. 2015;14(8):3174–87.
Wesseling H, Rahmoune H, Tricklebank M, Guest PC, Bahn S. A targeted multiplexed proteomic investigation identifies ketamine-induced changes in immune markers in rat serum and expression changes in protein kinases/phosphatases in rat brain. J Proteome Res. 2015;14(1):411–21.
Wesseling H, Guest PC, Lee CM, Wong EHF, Rahmoune H, Bahn S. Integrative proteomic analysis of the NMDA NR1 knockdown mouse model reveals effects on central and peripheral pathways associated with schizophrenia and autism spectrum disorders. Mol Autism. 2014;5:38.
Gottschalk MG, Wesseling H, Guest PC, Bahn S. Proteomic enrichment analysis of psychotic and affective disorders reveals common signatures in presynaptic glutamatergic signaling and energy metabolism. Int J Neuropsychopharmacol. 2014;18(2).
Clough T, Thaminy S, Ragg S, Aebersold R, Vitek O. Statistical protein quantification and significance analysis in label-free LC-MS experiments with complex designs. BMC Bioinformatics. 2012;13 Suppl 16:S6.
Wesseling H, Gottschalk MG, Bahn S. Targeted multiplexed selected reaction monitoring analysis evaluates protein expression changes of molecular risk factors for major psychiatric disorders. Int J Neuropsychopharmacol. 2014;18(1).
Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222.
Simicevic J, Schmid AW, Gilardoni PA, Zoller B, Raghav SK, Krier I, Gubelmann C, Lisacek F, Naef F, Moniatte M, et al. Absolute quantification of transcription factors during cellular differentiation using multiplexed targeted proteomics. Nat Methods. 2013;10(6):570–6.
Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9(6):555–66.
Method of the Year 2012. Nat Methods. 2013;10(1):1.
Aebersold R, Burlingame AL, Bradshaw RA. Western blots versus selected reaction monitoring assays: time to turn the tables? Mol Cell Proteomics. 2013;12(9):2381–2.
Rubin RD, Watson PD, Duff MC, Cohen NJ. The role of the hippocampus in flexible cognition and social behavior. Front Hum Neurosci. 2014;8:742.
Beadle JN, Tranel D, Cohen NJ, Duff MC. Empathy in hippocampal amnesia. Front Psychol. 2013;4:69.
Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9(5):341–55.
Happe F, Ronald A. The 'fractionable autism triad': a review of evidence from behavioural, genetic, cognitive and neural research. Neuropsychol Rev. 2008;18(4):287–304.
Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11(7):490–502.
Iadevaia V, Huo YL, Zhang Z, Foster LJ, Proud CG. Roles of the mammalian target of rapamycin, mTOR, in controlling ribosome biogenesis and protein synthesis. Biochem Soc Trans. 2012;40:168–72.
Tyler WA, Gangoli N, Gokina P, Kim HA, Covey M, Levison SW, Wood TL. Activation of the mammalian target of rapamycin (mTOR) is essential for oligodendrocyte differentiation. J Neurosci. 2009;29(19):6367–78.
Lebrun-Julien F, Bachmann L, Norrmen C, Trotzmuller M, Kofeler H, Ruegg MA, Hall MN, Suter U. Balanced mTORC1 activity in oligodendrocytes is required for accurate CNS myelination. J Neurosci. 2014;34(25):8432–48.
Lourenco T, Paes de Faria J, Bippes CA, Maia J, Lopes-da-Silva JA, Relvas JB, Graos M. Modulation of oligodendrocyte differentiation and maturation by combined biochemical and mechanical cues. Sci Rep. 2016;6:21563.
Wood TL, Bercury KK, Cifelli SE, Mursch LE, Min J, Dai J, Macklin WB. mTOR: a link from the extracellular milieu to transcriptional regulation of oligodendrocyte development. Asn Neuro. 2013;5(1):63–79.
Krishnan ML, Commowick O, Jeste SS, Weisenfeld N, Hans A, Gregas MC, Sahin M, Warfield SK. Diffusion features of white matter in tuberous sclerosis with tractography. Pediatr Neurol. 2010;42(2):101–6.
Liu J, Dietz K, DeLoyht JM, Pedre X, Kelkar D, Kaur J, Vialou V, Lobo MK, Dietz DM, Nestler EJ, et al. Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat Neurosci. 2012;15(12):1621-+.
Tkachev D, Mimmack ML, Huffaker SJ, Ryan M, Bahn S. Further evidence for altered myelin biosynthesis and glutamatergic dysfunction in schizophrenia. Int J Neuropsychopharmacol. 2007;10(4):557–63.
Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, Starkey M, Webster MJ, Yolken RH, Bahn S. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362(9386):798–805.
Zeidan-Chulia F, de Oliveira BN, Casanova MF, Casanova EL, Noda M, Salmina AB, Verkhratsky A. Up-regulation of oligodendrocyte lineage markers in the cerebellum of autistic patients: evidence from network analysis of gene expression. Mol Neurobiol. 2015.
Broek JAC, Lin Z, de Gruiter HM, van 't Spijker H, Haasdijk ED, Cox D, Ozcan S, van Cappellen GWA, Houtsmuller AB, Willemsen R, de Zeeuw CI, Bahn S. Synaptic vesicle dynamic changes in a model of fragile X. Mol Autism. 2016;7:17.
Bloemen OJ, Deeley Q, Sundram F, Daly EM, Barker GJ, Jones DK, van Amelsvoort TA, Schmitz N, Robertson D, Murphy KC, et al. White matter integrity in Asperger syndrome: a preliminary diffusion tensor magnetic resonance imaging study in adults. Autism Res. 2010;3(5):203–13.
Flores AI, Narayanan SP, Morse EN, Shick HE, Yin X, Kidd G, Avila RL, Kirschner DA, Macklin WB. Constitutively active Akt induces enhanced myelination in the CNS. J Neurosci. 2008;28(28):7174–83.
Narayanan SP, Flores AI, Wang F, Macklin WB. Akt signals through the mammalian target of rapamycin pathway to regulate CNS myelination. J Neurosci. 2009;29(21):6860–70.
Levi S, Logan SM, Tovar KR, Craig AM. Gephyrin is critical for glycine receptor clustering but not for the formation of functional GABAergic synapses in hippocampal neurons. J Neurosci. 2004;24(1):207–17.
Labonne JD, Graves TD, Shen Y, Jones JR, Kong IK, Layman LC, Kim HG. A microdeletion at Xq22.2 implicates a glycine receptor GLRA4 involved in intellectual disability, behavioral problems and craniofacial anomalies. BMC Neurol. 2016;16:132.
Wuerfel E, Bien CG, Vincent A, Woodhall M, Brockmann K. Glycine receptor antibodies in a boy with focal epilepsy and episodic behavioral disorder. J Neurol Sci. 2014;343(1–2):180–2.
Turner MR, Irani SR, Leite MI, Nithi K, Vincent A, Ansorge O. Progressive encephalomyelitis with rigidity and myoclonus Glycine and NMDA receptor antibodies. Neurology. 2011;77(5):439–43.
Harvey RJ, Yee BK. Glycine transporters as novel therapeutic targets in schizophrenia, alcohol dependence and pain. Nat Rev Drug Discov. 2013;12(11):866–85.
Jacobsson G, Meister B. Molecular components of the exocytotic machinery in the rat pituitary gland. Endocrinology. 1996;137(12):5344–56.
Whiteheart SW, Rossnagel K, Buhrow SA, Brunner M, Jaenicke R, Rothman JE. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J Cell Biol. 1994;126(4):945–54.
Yan Q, Sun W, McNew JA, Vida TA, Bean AJ. Ca2+ and N-ethylmaleimide-sensitive factor differentially regulate disassembly of SNARE complexes on early endosomes. J Biol Chem. 2004;279(18):18270–6.
Fernandez-Castillo N, Cormand B, Roncero C, Sanchez-Mora C, Grau-Lopez L, Gonzalvo B, Miquel L, Corominas R, Ramos-Quiroga JA, Casas M, et al. Candidate pathway association study in cocaine dependence: the control of neurotransmitter release. World J Biol Psychiatry. 2012;13(2):126–34.
Imai C, Sugai T, Iritani S, Niizato K, Nakamura R, Makifuchi T, Kakita A, Takahashi H, Nawa H. A quantitative study on the expression of synapsin II and N-ethylmaleimide-sensitive fusion protein in schizophrenic patients. Neurosci Lett. 2001;305(3):185–8.
Nishimune A, Isaac JT, Molnar E, Noel J, Nash SR, Tagaya M, Collingridge GL, Nakanishi S, Henley JM. NSF binding to GluR2 regulates synaptic transmission. Neuron. 1998;21(1):87–97.
Osten P, Srivastava S, Inman GJ, Vilim FS, Khatri L, Lee LM, States BA, Einheber S, Milner TA, Hanson PI, et al. The AMPA receptor GluR2 C terminus can mediate a reversible, ATP-dependent interaction with NSF and alpha- and beta-SNAPs. Neuron. 1998;21(1):99–110.
Cong M, Perry SJ, Hu LA, Hanson PI, Claing A, Lefkowitz RJ. Binding of the beta2 adrenergic receptor to N-ethylmaleimide-sensitive factor regulates receptor recycling. J Biol Chem. 2001;276(48):45145–52.
Heydorn A, Sondergaard BP, Hadrup N, Holst B, Haft CR, Schwartz TW. Distinct in vitro interaction pattern of dopamine receptor subtypes with adaptor proteins involved in post-endocytotic receptor targeting. FEBS Lett. 2004;556(1–3):276–80.
Bomberger JM, Parameswaran N, Hall CS, Aiyar N, Spielman WS. Novel function for receptor activity-modifying proteins (RAMPs) in post-endocytic receptor trafficking. J Biol Chem. 2005;280(10):9297–307.
Pontier SM, Lahaie N, Ginham R, St-Gelais F, Bonin H, Bell DJ, Flynn H, Trudeau LE, McIlhinney J, White JH, et al. Coordinated action of NSF and PKC regulates GABAB receptor signaling efficacy. EMBO J. 2006;25(12):2698–709.
Zheng Z, Keifer J. Protein kinase C-dependent and independent signaling pathways regulate synaptic GluR1 and GluR4 AMPAR subunits during in vitro classical conditioning. Neuroscience. 2008;156(4):872–84.
Gomes AR, Correia SS, Esteban JA, Duarte CB, Carvalho AL. PKC anchoring to GluR4 AMPA receptor subunit modulates PKC-driven receptor phosphorylation and surface expression. Traffic. 2007;8(3):259–69.
Silverman JL, Smith DG, Rizzo SJ, Karras MN, Turner SM, Tolu SS, Bryce DK, Smith DL, Fonseca K, Ring RH, et al. Negative allosteric modulation of the mGluR5 receptor reduces repetitive behaviors and rescues social deficits in mouse models of autism. Sci Transl Med. 2012;4(131):131ra151.
Berry-Kravis E, Des Portes V, Hagerman R, Jacquemont S, Charles P, Visootsak J, Brinkman M, Rerat K, Koumaras B, Zhu L, et al. Mavoglurant in fragile X syndrome: results of two randomized, double-blind, placebo-controlled trials. Sci Transl Med. 2016;8(321):321ra325.
Jacquemont S, Berry-Kravis E, Hagerman R, von Raison F, Gasparini F, Apostol G, Ufer M, Des Portes V, Gomez-Mancilla B. The challenges of clinical trials in fragile X syndrome. Psychopharmacology. 2014;231(6):1237–50.
Li NX, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, Li XY, Aghajanian G, Duman RS. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329(5994):959–64.
Huang LN, Liu Y, Jin W, Ji XC, Dong ZM. Ketamine potentiates hippocampal neurodegeneration and persistent learning and memory impairment through the PKC gamma-ERK signaling pathway in the developing brain. Brain Res. 2012;1476:164–71.
Shapira M, Kakiashvili E, Rosenberg T, Hershko DD. The mTOR inhibitor rapamycin down-regulates the expression of the ubiquitin ligase subunit Skp2 in breast cancer cells. Breast Cancer Res. 2006;8(4):R46.
Reinhard JR, Kriz A, Galic M, Angliker N, Rajalu M, Vogt KE, Ruegg MA. The calcium sensor Copine-6 regulates spine structural plasticity and learning and memory. Nat Commun. 2016;7:11613.
Suetsugu M, Mehraein P. Spine distribution along the apical dendrites of the pyramidal neurons in Down's syndrome. A quantitative Golgi study. Acta Neuropathol. 1980;50(3):207–10.
Chang KT, Ro H, Wang W, Min KT. Meeting at the crossroads: common mechanisms in Fragile X and Down syndrome. Trends Neurosci. 2013;36(12):685–94.
Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10(10):981–91.
Swann JW, Al-Noori S, Jiang M, Lee CL. Spine loss and other dendritic abnormalities in epilepsy. Hippocampus. 2000;10(5):617–25.
Chang CY, Picotti P, Huttenhain R, Heinzelmann-Schwarz V, Jovanovic M, Aebersold R, Vitek O. Protein significance analysis in selected reaction monitoring (SRM) measurements. Mol Cell Proteomics. 2011;11(4):M111 014662.
Benjamini Y, Hochberg Y. Controlling the false discovery rate— a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B Methodol. 1995;57(1):289–300.
Acknowledgements
We would like to thank Susan Gooden for the generation and treatment of the Tsc1 +/- mouse model and Jadviga Schreiber for proofreading the manuscript.
Funding
This research was kindly supported by the Stanley Medical Research Institute (SMRI) and the Dutch Fund for Economic Structure Reinforcement (#0908) the NeuroBasic PharmaPhenomics project.
Availability of data and materials
The datasets presented in this study can be made available on reasonable request.
Author information
Authors and Affiliations
Contributions
HW carried out the label-free LC-MSE experiments, designed and carried out the SRM experiments and performed all statistical and bioinformatic data analyses. HW prepared the figures and tables and drafted the manuscript. SB and YE conceived the study and participated in its design and coordination. SB helped to interpret the results and to write the manuscript. All authors read, edited and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All animal experiments were approved by the Dutch Ethical Committee or in accordance with Institutional Animal Care and Use Committee guidelines.
All animal experiments were approved by the Dutch Animal Experiment Committee (Dierexperimenten commissie [DEC]) and in accordance with Dutch animal care and use laws.
Consent for publication
Not applicable.
Competing interests
S.B. is a director of Psynova Neurotech Ltd. and PsyOmics Ltd. The other authors declare no conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Supplementary methods. Detailed information of the experimental methods. (DOCX 31 kb)
Additional file 2:
Significantly altered proteins identified by label-free LC-MSE analysis in the frontal cortex and hippocampus of the Tsc1 +/− mice compared to Wt mice. Overlapping proteins between hippocampus and frontal cortex are bold. (XLSX 23 kb)
Additional file 3:
Full information for significantly changed proteins identified by label-based LC-SRM in the frontal cortex and hippocampus of Tsc1 +/− mice compared to wildtype mice. (DOCX 31 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wesseling, H., Elgersma, Y. & Bahn, S. A brain proteomic investigation of rapamycin effects in the Tsc1 +/− mouse model. Molecular Autism 8, 41 (2017). https://doi.org/10.1186/s13229-017-0151-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13229-017-0151-y