Abstract
Background
Vibrio parahaemolyticus is a causative agent of gastroenteritis. Most of the clinical isolates carry either tdh and/or trh genes which are considered as the major virulence genes of this pathogen. In this study, the clinical isolates of V. parahaemolyticus carrying trh gene (n = 73) obtained from 1886 to 2012 from various countries were investigated for the urease production, haemolytic activity, and biofilm formation. In addition, the potential of clustered regularly interspaced short palindromic repeats (CRISPR)-based genotyping among these isolates was investigated.
Results
In this study, no significant differences were observed in the urease production between tdh+ trh1+ and tdh+ trh2+ isolates (p = 0.063) and between the tdh− trh1+ and tdh− trh2+ isolates (p = 0.788). The isolates carrying only the trh gene showed variation in their haemolytic activity. The ratio of urease production and haemolytic activity between the trh1+ and trh2+ isolates and biofilm formation of trh+ V. parahaemolyticus isolates were not significantly different. Sixteen of thirty-four tested isolates (47.0%) of trh+ V. parahaemolyticus were positive for CRISPR detection. The discriminatory power index (DI) of CRISPR-virulence typing was higher than the DI obtained by CRISPR typing alone.
Conclusion
The tdh and trh genes were not involved in urease production in the trh+ V. parahaemolyticus, and variation of haemolytic activity detected in V. parahaemolyticus carrying only the trh gene might be correlated to the sequence variation within trh1 and trh2 genes. Additionally, biofilm production of V. parahaemolyticus was not associated with harboring of virulence genes. For genotyping, CRISPR sequences combined with virulence genes can be used as genetic markers to differentiate trh+ V. parahaemolyticus strains.
Similar content being viewed by others
Background
Vibrio parahaemolyticus is a halophilic Gram-negative bacterium that occurs naturally in worldwide aquatic environments. The organism causes gastroenteritis in several countries due to consumption of raw or undercooked seafood [1]. The pathogenicity of this bacterium in humans is associated to the production of thermostable direct hemolysin (TDH) encoded by tdh gene and/or TDH-related hemolysin (TRH) encoded by trh gene, which are considered to be the major virulence factors, however, not many isolates from the environment possess these genes [2]. V. parahaemolyticus obtained the tdh and trh genes from other organisms and transmit them among V. parahaemolyticus strains via horizontal gene transfer [3]. TDH and TRH are approximately 67% identity in their amino acid sequences and possess common biological activities including haemolytic activity, enterotoxicity, cytotoxicity, and cardiotoxicity [4]. The trh gene possesses a significantly broader nucleotide sequence variation and can be subdivided into two main subtypes (trh1 and trh2) which share 84% identity in their sequences [5]. The presence of trh gene in V. parahaemolyticus isolates is associated with the urease production because of the genetic linkage between ure and trh genes on the chromosome of trh+ V. parahaemolyticus [6]. Urease is an enzyme that catalyzes the hydrolysis of urea to ammonia, raising the pH of environment inside the host. It is possible that urease contributes to the pathogenicity of this bacterium by increasing its chance of survival after entering the human body, as has been previously demonstrated for Yersinia enterocolitica infections [7]. Clustered regularly interspaced short palindromic repeats (CRISPR) and repetitive sequences such as tandem repeats are commonly detected in the genomes of bacteria. Their lengths and numbers are highly variable among bacterial strains and are useful for bacterial genotyping [8]. CRISPR is a segment of prokaryotic DNA containing direct repeats (DRs), typically 24–47 bp nucleotides in length, and spacer (non-repetitive sequences obtained from foreign genetic elements). The polymorphism detected in CRISPR loci has been applied as a genetic marker for typing many bacteria, such as Campylobacter fetus and S. Typhimurium [9, 10]. Association of CRISPR and virulence factors of bacteria has been demonstrated in Pseudomonas aeruginosa, Myxococcus xanthus, Francisella novicida and Listeria monocytogenes [11]. In V. parahaemolyticus, correlation between CRISPR and virulence genes was determined in 208 isolates from clinical and food samples [12]. CRISPR was detected in 149 of 153 (97.4%) of tdh+ isolates. In contrast, among the 6 trh+ V. parahaemolyticus tested isolates, only 2 were positive for CRSIPR locus [12]. Therefore, association of CRISPR and the trh+ V. parahaemolyticus is not completely concluded.
In this work, trh+ V. parahaemolyticus isolates from clinical samples were investigated for urease production, haemolytic activity and biofilm formation. In addition, molecular typing based on CRISPR was analyzed.
Methods
Bacterial strains
A total of 73 clinical trh+ V. parahaemolyticus isolates were obtained from various countries between 1886 and 2012 (Table 1). They belonged to 28 different serotypes consisting of 10 O serogroups and 19 K antigens. All isolates were confirmed as V. parahaemolyticus using PCR targeted to the toxR gene [13].
Detection of virulence genes
Genomic DNA of all V. parahaemolyticus isolates was extracted using boiling method [14] and was used as templates to detect the virulence genes, tdh, trh1 and trh2. A 251-bp sequence of tdh gene was amplified by PCR [15]. For detection of trh genes, PCR was performed using two primer sets based on trh1 and trh2 sequences available in the NCBI GenBank database. The trh1 primers: Trh1-F1 (5ʹ-CTGAATCACCAGTTAACGC-3ʹ) and Trh1-R1: (5ʹ-GGCGTTTRATCCAAATAC-3ʹ) generated a PCR product of 313-bp and trh2 primers: Trh2-F2 (5ʹ-CAATCAAAACTGAATCCCC-3ʹ) and Trh2-R3 (5ʹ-CATCAACAAAAMATTTTACCGA-3ʹ) provided an amplicon of 276-bp. The PCR reaction was carried out with a reaction mixture consisting of 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 μM of each primer, 0.025 U of GoTaq DNA polymerase and 2.0 μl DNA templates in a 20 μl volume. The reactions were performed with a Thermal Cycler Gene Atlas (Astec, Fukuoka, Japan) as follows: 5 min for a hot start at 96 °C, followed by 35 cycles of amplification consisting of denaturation at 94 °C for 1 min, annealing at 53 °C for 1 min and extension at 72 °C for 1 min and final extension at 72 °C for 7 min. Electrophoresis was performed on a 1.5% agarose gel and the amplicons were detected using a UV transilluminator. In addition, the specificity of the trh1 and trh2 primers was also determined by purification of PCR products using ethanol/sodium acetate precipitation, and sequencing.
Quantitative urease assay
Urease was quantified using a colorimetric assay based on the reaction of ammonia (NH3) and phenol in the presence of hypochlorite which yields a blue product of indophenol [16]. Briefly, bacteria were inoculated in LB broth supplemented with 3% sodium chloride (NaCl) and 0.1% urea and incubated at 37 °C. Overnight cultures were concentrated by centrifugation, washed twice with 50 mM HEPES buffer (pH 7.5) and resuspended in the same buffer. The cells were lysed by sonication, then, 50 μl of supernatant obtained after centrifugation was mixed with 25 mM of urea in HEPES buffer (pH 7.5). After incubation at 37 °C for 30 min, ammonia released from the lysate was determined by adding solution of 1% (w/v) phenol and 170 μM sodium nitroprusside followed by solution of 125 mM sodium hydroxide (NaOH) and 0.05% (w/v) sodium hypochlorite (NaOCl). Subsequently, the tubes were incubated at 37 °C for 30 min and the absorbance was determined on a Hitachi U 2000 Double-Beam UV/VIS spectrophotometer (Hitachi Instruments Inc., Danbury, CT) at a wavelength of 625 nm. Ammonium chloride (NH4Cl) was used to perform standard curve. The total protein concentration from the same lysate was determined by Lowry’s method with Folin-Ciocalteu’s reagent solution (Nacalai Tesque Inc., Kyoto, Japan) and bovine serum albumin (BSA) was used to set up standard curve [17]. Urease activity was calculated as micromoles of NH3 per minute per milligram of protein.
Determination of haemolytic activity
V. parahaemolyticus carrying either the trh1 or trh2 gene could lyse human erythrocyte [5, 18]. In this study, haemolytic activity of 15 tdh− trh1+ and 20 tdh− trh2+ isolates of V. parahaemolyticus was evaluated using the blood agarose assay [19] with some modification. Briefly, V. parahaemolyticus was grown in LB broth supplemented with 2% NaCl at 37 °C for 18 h, the pellet was harvested and resuspended in PBS (pH 7.0). Then, the cells were sonicated and the supernatant obtained by centrifugation was determined. The 50 μl of the supernatant was added into the agarose wells containing various concentration of human blood (1%, 0.5% and 0.25%). After incubation at 37 °C for 24 h, a clear zone around the well was indicated as haemolytic activity. High haemolytic activity (+3) was defined as the isolates can lyse all three concentration of erythrocytes whereas medium (+2) and low (+1) haemolytic activities were defined as the isolates can haemolyse two (0.5% and 0.25%) and one (0.25%) blood concentrations, respectively.
Quantitative biofilm assays
Each 5 isolates of V. parahaemolyticus belonging to the tdh− trh1+, tdh− trh2+, tdh+ trh1+ and tdh+ trh2+ isolates was investigated for biofilm formation as previously described [20]. Biofilm formation was quantified by measuring the optical density (OD) at 570 nm using a LUMIstar Omega spectrophotometer (BMG LABTECH, Germany). LB broth supplemented with 2% NaCl was used as control. The experiment was performed in triplicate.
Determination of CRISPR using PCR technique
The primers for detection of CRISPR of V. parahaemolyticus were designed from the CRISPR sequences of V. parahaemolyticus serotype O3:K6 (strain RIMD2210633) obtained from the CRISPR database website (http://crispr.u-psud.fr/) [21]. The forward primer: VpCRISPR_3-F (5ʹ-ATGCATTCCAAAGCTACCACTC-3ʹ) and the reverse primer: VpCRISPR_705-R (5ʹ-GCCTACCAGATAGCAAGTGTCC-3ʹ) generated a 592-bp product. The PCR reaction mixture consisted of 1 × Ex Taq Buffer, 2 mM MgCl2, 1.25 U Takara Ex Taq DNA polymerase (Takara Biochemicals, Tokyo, Japan), 200 μM dNTPs, 0.2 μM of each primer, and 5 μl of DNA template in a total volume of 50 μl. The PCR reaction was performed using the following conditions: initial denaturation cycle for 1 min at 94 °C, followed by 30 cycles of amplification consisting of denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min and extension at 72 °C for 1 min and final extension at 72 °C for 5 min. The PCR product was confirmed by sequencing using the forward primer: VpCRISPR_3-F.
CRISPR pattern including the DRs and spacers was investigated using the CRISPRfinder tool (http://crispr.i2bc.paris-saclay.fr/Server/). The DR sequences in each isolate were analyzed based on the similarity of consensus direct repeat sequences (CDRs). In addition, all spacer sequences were investigated using the CRISPRTarget tool (http://bioanalysis.otago.ac.nz/CRISPRTarget/) and were used for phylogenetic tree construction.
In this work, CRISPR-virulence typing was constructed and compared to profiles obtained by CRISPR typing alone. CRISPR-virulence typing was constructed based on the CRISPR spacer sequences and the presence of virulence genes that included the tdh, trh1 and trh2 genes. A profile of each isolate was created using a binary matrix of presence or absence of spacer sequences and virulence genes. The dendrogram was constructed using BioNumerics 7.0 software (Applied Maths, Saint-Martens-Latem, Belgium) with the UPGMA algorithm using the Dice similarity coefficient. The discriminatory power index (DI) of CRISPR typing alone and CRISPR-virulence typing were assessed by Simpson’s diversity index [22].
Statistical analysis
Urease activity and correlation between urease and haemolytic activities of trh+ V. parahaemolyticus isolates were determined by the independent samples t test analysis using SPSS 11.5 software. The differences observed were considered statistically significant at p < 0.05.
Results and discussion
Urease production in trh + V. parahaemolyticus
Urease is detected in many pathogenic bacteria such as Proteus mirabilis, Helicobacter pylori and Yersinia enterocolitica [23,24,25]. In V. parahaemolyticus, urease-positive phenotype strongly correlates with the possession of the trh gene making it as a marker of trh+ strains [26]. In this work, urease production in trh+ V. parahaemolyticus isolates was evaluated. Previous results have suggested that the presence of urea is involved in the urease induction of trh+ V. parahaemolyticus TH3996 strain [27]. Therefore, in this work, all tested isolates were grown in the liquid medium supplemented with 0.1% urea before testing. Urease production of the tdh+ trh1+, tdh+ trh2+, tdh− trh1+and tdh− trh2+ V. parahaemolyticus isolates varied from 0.46 to 15.54, 1.75 to 21.05, 0.89 to 20.79 and 3.46 to 21.20 μmol NH3/min/mg protein, respectively. No significant differences were observed in the urease production between the tdh+ trh1+ and tdh+ trh2+ (p = 0.063) as well as in the tdh− trh1+ and tdh− trh2+ isolates (p = 0.788) (Fig. 1). These indicated that urease production among clinical trh+ V. parahaemolyticus isolates varied distinctively but it was not associated with either the trh or tdh genes. In addition, no correlation between urease production and serotypes of V. parahaemolyticus was observed in this study (Additional file 1: Table S1).
Comparison of urease production between trh+ V. parahaemolyticus isolates according to the presence or absence of tdh. Horizontal bar within box represents median values and vertical line out of the box indicates minimum and maximum. The difference in urease production in each group was compared using the independent samples t-test analysis
Haemolytic activity of trh + V. parahaemolyticus
A total of 15 trh1+ isolates was determined, 13.3%, 53.3% and 33.3% of the total isolates exhibited high, medium and low haemolytic activity, respectively (Fig. 2). For 20 isolates of V. parahaemolyticus carrying the trh2 gene, 5%, 35% and 60% of the total isolates displayed high, medium and low haemolytic activity. It has been demonstrated that the expression of trh2 was lower than the trh1 [5, 28]. Therefore, the results obtained in this study might correlate to the expression of the genes and the sequences variation within the trh1 and trh2 genes [5].
In this work, correlation between urease production and haemolytic activity of trh+ isolates was evaluated. Although the urease production and haemolytic activity ratio of trh2+ isolates was higher (1.65 to 21.20 μmol NH3/min/mg protein) than that of the trh1+ isolates (0.56–19.21 μmol NH3/min/mg protein), no significant difference was observed (p = 0.683) (Fig. 3). This indicated the urease was not involved in the human erythrocyte lysis by TRH hemolysin of trh+ V. parahaemolyticus strains.
Correlation between urease production and haemolytic activity of V. parahaemolyticus isolates carrying only the trh gene. Horizontal bar within box represents median values and vertical lines out of the box indicate minimum and maximum. ◯ indicates the outlier. No significant differences were observed in the urease production/haemolytic activity between the isolates carrying the trh1 and trh2 genes (p = 0.683) using the independent samples t-test analysis
Biofilm formation
Most of human bacterial infections are associated with biofilms that contributed to their resistance and persistence in the host [29]. Previous study demonstrated that biofilm formation of vibrios was a survival mechanism associated with their pathogenesis and stress tolerance [30]. In order to investigate the involvement of the virulence genes and biofilm production, 4 categories of V. parahaemolyticus (tdh+ trh1+, tdh+ trh2+, tdh− trh1+, and tdh− trh2+) were determined. There was no any difference in biofilm formation among 4 categories of V. parahaemolyticus because high variation of biofilm formation within the isolates in each category was observed (Fig. 4). Variation in biofilm formation among the bacterial isolates has been reported. High variability in biofilm formation of 34 strains of Acinetobacter baumannii isolated from hospitalized patients was observed without correlation to molecular types and antimicrobial resistance [31]. Ninety-eight strains with the same serotype of Listeria monocytogenes displayed different biofilm formation [32]. Additionally, quantitative biofilm assay of Vibrio cholerae isolates in Thailand demonstrated that those isolates possessed a wide range of biofilm production [33].
CRISPR sequences in trh + V. parahaemolyticus
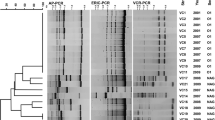
In the present study, specific primer pairs for detecting CRISPR region were designed and compared to the previously reported by Sun and co-workers [12]. No difference in the specificity detected in the 5 V. parahaemolyticus tested isolates after confirmation by sequencing (data not shown). Thirty-four isolates of trh+ V. parahaemolyticus including 10 isolates of tdh+ trh1+ and each 8 isolates of tdh+ trh2+, tdh− trh1+, tdh− trh2+ were selected for CRISPR sequences analysis. Sixteen isolates (47.0%) were positive for CRISPR, they were 8, 5 and 3 from tdh+ trh1+, tdh+ trh2+, and tdh− trh2+ isolates, respectively (Table 2). None of the isolate carrying only the trh1 gene was positive for CRISPR. The result indicated that the number of CRISPR-positive isolates of trh+ V. parahaemolyticus obtained in this study was higher than the 2 out of 6 (33.3%) isolates reported in a previous study [12]. It is of interest that CRISPR was mostly detected in the isolates carrying the trh gene together with the tdh gene. Association of tdh and the presence of CRISPR was observed in 97.4% of V. parahaemolyticus isolates [12]. Thus, horizontal gene transfer of virulence genes might have occurred among V. parahaemolyticus and close related species. The numbers of DRs were between 25 and 28 bp nucleotides in lengths and the DR unique sequences of all CRISPR-positive isolates were GTGAACTGCCGAATAGGTAGCTGAT (Table 2). A total of 28 spacers were obtained and the number of spacer detected in each isolate was between 1 or 2 with 30 to 32 bp nucleotides in lengths (Additional file 1: Table S1). Spacer analysis using the CRISPRTarget and the BLAST databases from NCBI revealed that most of them showed 87–100% similarity to Vibrio alginolyticus plasmids except one spacer of PSU5256 suggesting the possibility of genetic transfer between V. alginolyticus and V. parahaemolyticus (Additional file 2: Table S2). Phylogenetic tree of all 28 spacers were generated and 6 different spacer patterns designated as SP1 to SP6 were classified using maximum likelihood method (Fig. 5).
Phylogenetic relationship of all 28 spacer sequences detected in 16 CRISPR-positive V. parahaemolyticus isolates. The tree was constructed using maximum likelihood method. Numbers at branch-points represent confidence values obtained after bootstrap analysis of the maximum likelihood tree using 1000 replicates. The scale bar represents 0.1 substitutions per nucleotide position. PSU5256-s2, spacer no. 2 of V. parahaemolyticus PSU5256; C1, tdh+ trh1+; C2, tdh+ trh2+; C3, tdh− trh1+; C4, tdh− trh2+
CRISPR analysis based on the CRISPR spacer sequences has been applied for bacterial subtyping such as Campylobacter jejuni, Mycobacterium tuberculosis, Salmonella enterica and Yersinia pestis [9, 34,35,36]. In this work, a total of 16 of 34 trh+ V. parahaemolyticus isolates were positive for CRISPR detection, and they were classified into 5 CRISPR types (CTs) based on CRISPR spacer patterns at 75% similarity level (Fig. 6). All isolates in CT1 type were trh1+ that contained spacers SP4 and SP6. CT2 of both the trh1+ and trh2+ isolates harbored spacers SP1 and SP6. One isolate of trh2+ V. parahaemolyticus containing spacers SP1 and SP5 was classified into CT3 type. However, CT4 and CT5 contained only one spacer (SP3 in CT4; SP2 in CT5) (Fig. 6). Although the tested V. parahaemolyticus isolates were classified using CPISPR-based typing, they could not be grouped according to the trh gene harboring (trh1 or trh2) (Fig. 6).
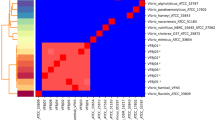
Previous study demonstrated that a combination of CRISPR and virulence genes significantly increased the discriminatory power and could be a useful subtyping method for investigation of Salmonella outbreaks [37]. In this work, a combination of CRISPR spacer sequences with virulence genes (tdh, trh1 and trh2 genes) of V. parahaemolyticus isolates was investigated. CRISPR-virulence typing profiles obtained from 34 isolates of trh+ V. parahaemolyticus were organized into 7 clusters with 12 different profiles at 75% similarity level (Fig. 7). The isolates within the same cluster possessed the identical subtype of the trh gene (either trh1 or trh2). All trh1+ isolates were classified in the CV1, CV2 and CV3 clusters of CRISPR-virulence typing, while the remaining four clusters (CV4 to CV7) were trh2+ isolates. The isolates in clusters CV1, CV2, CV4 and CV6 possessed the tdh gene, but not all of them gave positive results for CRISPR detection. The CV3 and CV5 clusters were negative for CRISPR (Fig. 7). Identical CRISPR-virulence typing profiles were detected in clusters CV1 (PSU5105 and PSU5106; PSU4921, PSU5107, PSU5296 and PSU5322), CV2 (PSU5069 and PSU5305; PSU5264 and 1884) and CV6 (PSU5323 and PSU5331; 2435, 2443 and 2463) (Fig. 7). It is postulated that these bacteria might obtain the plasmids or exogenous genetic elements derived from the same origin. It was of interest that the spacers SP2 and SP5 were not detected in trh1+ isolates, whereas trh2+ isolates lacked spacer SP4 (Fig. 7). The spacer SP2 and SP4 were homologous to V. alginolyticus plasmids, whereas spacer SP5 showed no sequence homology with any bacteriophages or plasmids (Additional file 2: Table S2). Self-derived spacers have been detected in some microbial genomes [38,39,40]. It has been postulated that some bacterial spacers may be self-targeting spacers that are a form of autoimmunity [40, 41].
The discriminatory power index (DI) of both CRISPR analysis and CRISPR-virulence typing were evaluated. DI of CRISPR-virulence typing (DI = 0.90) was higher than that of CRISPR typing (DI = 0.67). This was in agreement with a recent study that DI of CRISPR-virulence typing of Helicobacter pylori was higher than CRISPR typing alone [42]. Although, pulse field gel electrophoresis (PFGE) is a good method for discrimination of V. parahaemolyticus, it is laborious, time consuming [43, 44]. In this study, Analysis of CRISPR spacers and virulence genes were evaluated for typing trh+ V. parahaemolyticus. CRISPR-virulence gene typing is a method based on PCR detection that provided high DI (0.90). It is easy to perform which required only PCR machine, therefore, CRISPR-virulence gene typing could be a useful method for typing trh+ V. parahaemolyticus.
Conclusions
A total of 73 isolates of trh+ V. parahaemolyticus obtained from clinical samples were characterized. There was no significant difference in the urease production between the tdh+ trh1+ and tdh+ trh2+ and between the tdh− trh1+ and tdh− trh2+ isolates indicating that the tdh and trh genes were not involved in urease production in the trh+ V. parahaemolyticus isolates. The haemolytic activity of trh1+ isolates was higher than the trh2+ isolates. Variation in biofilm production was detected in the isolates belonging to the tdh+ trh1+, tdh+ trh2+, tdh− trh1+, and tdh− trh2+ groups. For genotyping, combination of CRISPR spacers and virulence genes provide high discriminatory power than that of CRISPR typing alone and it was able to distinguish between trh1+ and trh2+ V. parahaemolyticus isolates. Thus, CRISPR-virulence gene typing can be a useful method for typing trh+ V. parahaemolyticus strains.
References
Chiou CS, Hsu SY, Chiu SI, Wang TK, Chao CS. Vibrio parahaemolyticus serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J Clin Microbiol. 2000;38:4621–5.
Vuddhakul V, Chowdhury A, Laohaprertthisan V, Pungrasamee P, Patararungrong N, Thianmontri P, Ishibashi M, Matsumoto C, Nishibuchi M. Isolation of a pandemic O3:K6 clone of a Vibrio parahaemolyticus strain from environmental and clinical sources in Thailand. Appl Environ Microbiol. 2000;66:2685–9.
Terai A, Baba K, Shirai H, Yoshida O, Takeda Y, Nishibuchi M. Evidence for insertion sequence-mediated spread of the thermostable direct hemolysin gene among Vibrio species. J Bacteriol. 1991;173:5036–46.
Nishibuchi M, Fasano A, Russell RG, Kaper JB. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect Immun. 1992;60:3539–45.
Kishishita M, Matsuoka N, Kumagai K, Yamasaki S, Takeda Y, Nishibuchi M. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl Environ Microbiol. 1992;58:2449–57.
Iida T, Park KS, Suthienkul O, Kozawa J, Yamaichi T, Yamamoto K, Honda T. Close proximity of the tdh, trh, and ure genes on the chromosome of Vibrio parahaemolyticus. Microbiology. 1998;144:2517–23.
De Koning-Ward TF, Robins-Browne RM. Contribution of urease to acid tolerance in Yersinia enterocolitica. Infect Immun. 1995;3:3790–5.
Lupski JR, Roth JR, Weinstock GM. Chromosomal duplications in bacteria, fruit flies, and humans. Am J Hum Genet. 1996;58:21–7.
Calleros L, Betancor L, Iraola G, Méndez A, Morsella C, Paolicchi F, Silveyra S, Velilla A, Pérez R. Assessing the intra–species genetic variability in the clonal pathogen Campylobacter fetus: CRISPRs are highly polymorphic DNA markers. J Microbiol Methods. 2017;132:86–94.
Almeida F, Medeiros MI, Rodrigues DD, Allard MW, Falcao JP. Molecular characterization of Salmonella typhimurium isolated in Brazil by CRISPR-MVLST. J Microbiol Methods. 2017;133:55–61.
Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol. 2014;12:317–26.
Sun H, Li Y, Shi X, Lin Y, Qiu Y, Zhang J, Liu Y, Jiang M, Zhang Z, Chen Q, Sun Q, Hu Q. Association of CRISPR/Cas evolution with Vibrio parahaemolyticus virulence factors and genotypes. Foodborne Pathog Dis. 2015;12:68–73.
Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1999;37:1173–7.
Theron J, Cilliers J. Du Preez M, Bro È zel1 VS, Venter1 SN. Detection of toxigenic Vibrio cholerae from environmental water samples by an enrichment broth cultivation-pit-stop semi-nested PCR procedure. J Appl Microbiol. 2000;89:539–46.
Tada J, Ohashi T, Nishimura N, Shirasaki Y, Ozaki H, Fukushima S, Takano J, Nishibuchi M, Takeda Y. Detection of thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Probes. 1992;6:477–87.
Weatherburn MW. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–4.
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75.
Honda T, Ni Y, Miwatani T. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect Immun. 1988;56:961–5.
Nishibuchi M, Kaper JB. Nucleotide sequence of the thermostable direct hemolysin gene of Vibrio parahaemolyticus. J Bacteriol. 1985;162:558–64.
Kongrueng J, Mitraparp-Arthorn P, Bangpanwimon K, Robins W, Vuddhakul V, Mekalanos J. Isolation of Bdellovibrio and like organisms and potential to reduce acute hepatopancreatic necrosis disease caused by Vibrio parahaemolyticus. Dis Aquat Org. 2017;124:223–32.
Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 2007;8:172.
Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–6.
Probst P, Hermann E, zum Büschenfelde KM, Fleischer B. Zum Buschenfelde KH, Fleischer B. Identification of the Yersinia enterocolitica urease subunit as a target antigen for human synovial T lymphocytes in reactive arthritis. Infect Immun. 1993;61:4507–9.
Sriwanthana B, Island MD, Mobley HLT. Sequence of the Proteus mirabilis urease accessory gene ureG. Gene. 1993;129:103–6.
Voland P, Weeks DL, Marcus EA, Prinz C, Sachs G, Scott D. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am J Physiol Gastrointest Liver Physiol. 2003;284:96–106.
Suthienkul O, Ihibashi M, Iida T, Nettip N, Supavej S, Eampokalap B, Makino M, Honda T. Urease production correlates with possession of the trh gene in Vibrio parahaemolyticus strains isolated in Thailand. J Infect Dis. 1995;172:1405–8.
Park KS, Iida T, Yamaichi Y, Oyagi T, Yamamoto K, Honda T. Genetic characterization of DNA region containing the trh and ure genes of Vibrio parahaemolyticus. Infect Immun. 2000;68:5742–8.
Okuda J, Nishibuch M. Manifestation of the Kanagawa phenomenon, the virulence-associated phenotype, of Vibrio parahaemolyticus depend on a particular single base change in the promoter of the thermostable direct hemolysin. Mol Microbiol. 1998;39:499–511.
Abdel-Aziz SM, Aeron A. Bacterial biofilm: dispersal and inhibition strategies. SAJ Biotechnol. 2014;1:1–10.
Yildiz FH, Visic KV. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 2009;17:109–18.
Wroblewska MM, Sawicka-Grzelak A, Marchel H, Luczak M, Sivan A. Biofilm production by clinical strains of Acinetobacter baumannii isolated from patients hospitalized in two tertiary care hospitals. FEMS Immunol Med Microbiol. 2008;53:140–4.
Doijad SP, Barbuddhe SB, Garg S, Poharkar KV, Kalorey DR, Kurkure NV, Rawool DB, Chakraborty T. Biofilm-forming abilities of Listeria monocytogenes serotypes isolated from different sources. PLoS ONE. 2015;10:e0137046.
Preeprem S, Mittraparp-arthorn P, Bhoopong P, Vuddhakul V. Isolation and characterization of Vibrio cholerae isolates from seafood in Hat Yai City, Songkhla, Thailand. Foodborne Pathog Dis. 2014;11:881–6.
Fricke W, Mammel M, McDermott P, Tartera C, White D, Leclerc J, Ravel J, Cebula T. Comparative genomics of 28 Salmonella enteric isolates: evidence for CRISPR-mediated adaptive sublineage evolution. J Bacteriol. 2011;193:3556–68.
Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–63.
Shariat N, Dudley EG. CRISPRs: molecular signatures used for pathogen subtyping. Appl Environ Microbiol. 2014;80:430–9.
Liu F, Kariyawasam S, Jayarao BM, Barrangou R, GernerSmidt P, Ribot EM, Knabel SJ, Dudley EG. Subtyping Salmonella enterica serovar enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs). Appl Environ Microbiol. 2011;7:4520–6.
Hooton SP, Brathwaite KJ, Connerton IF. The bacteriophage carrier state of Campylobacter jejuni features changes in host non-coding RNAs and the acquisition of new host-derived CRISPR spacer sequences. Front Microbiol. 2016;7:355.
Hooton SP, Connerton IF. Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front Microbiol. 2014;5:744.
Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–40.
Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–82.
Bangpanwimon K, Sottisuporn J, Mittraparp-arthorn P, Ueaphatthanaphanich W, Rattanasupar A, Pourcel C, Vuddhakul C. CRISPR-like sequences in Helicobacter pylori and application in genotyping. Gut Pathog. 2017;9:65.
Kam KM, Luey CKY, Parsons MB, Cooper KLF, Nair GB, Alam M, Islam MA, Cheung DTL, Chu YW, Ramamurthy T, Pazhani GP, Bhattacharya SK, Watanabe H, Terajima J, Arakawa E, Ratchtrachenchai OA, Huttayananont S, Ribot EM, Gerner-Smidt P, Swaminathan B. Evaluation and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping Vibrio parahaemolyticus: an international multicenter collaborative study. J Clin Microbiol. 2008;46:2766–73.
Martinez-Urtaza J, Lozano-Leon A, DePaola A, Ishibashi M, Shimada K, Nishibuchi M, Liebana E. Characterization of pathogenic Vibrio parahaemolyticus isolates from clinical sources in Spain and comparison with Asian and North American pandemic isolates. J Clin Microbiol. 2004;42:4672–8.
Authors’ contributions
Conceptualization: VV, Formal analysis: PM, MN, KS. Investigation: JK, PM. Methodology: JK, PM, MN. Supervision: VV. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this published article (and its additional files).
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was financially supported by the Thailand Research Fund (TRF) through the Royal Golden Jubilee Ph.D. Program (Grant No. PHD/0213/2556) and the Center for Southeast Asia Studies (CSEAS), Kyoto University.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
Urease production, haemolytic activity and the ratio of urease production to haemolytic activity of all 73 trh+ V. parahaemolyticus isolates.
Additional file 2: Table S2.
Foreign genetic element similar to spacers using CRISPR targets analysis.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kongrueng, J., Srinitiwarawong, K., Nishibuchi, M. et al. Characterization and CRISPR-based genotyping of clinical trh-positive Vibrio parahaemolyticus. Gut Pathog 10, 48 (2018). https://doi.org/10.1186/s13099-018-0275-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13099-018-0275-4