Abstract
Background
Recent studies show that some Escherichia coli strains possessing a gene cluster named the pks island might have a causative role in the development of human colorectal cancer (CRC). In several reports from Europe, they are found more prevalently in colon tissue specimens derived from CRC patients compared to those from controls. In this study we sought to clarify the difference in pks prevalence between CRC patients and non-CRC controls in the Japanese population, by using non-invasive sample collection technique during colonoscopy.
Methods
Colonic lavage samples were collected during diagnostic colonoscopy, and bacterial DNA within each sample was extracted. Fecal DNA samples were then examined for pks island genes using conventional qualitative PCR and real-time quantitative PCR. In some patients biopsy samples were also collected in the same session of colonoscopy, and the correlation between the pks status of the colonic lavage sample and the biopsy sample of the same patients was evaluated.
Results
Twelve out of thirteen patients (92%) showed the same pks status by colonic lavage sample and biopsy sample, suggesting the usefulness of colonic lavage samples as a surrogate for biopsy samples. A total of 98 colonic lavage samples were collected, which included 35 from CRC patients, 37 from adenoma patients, and 26 from controls. The pks-positive bacterial DNA was detected in 43, 51, and 46% of colonic lavage samples from CRC, adenoma, and control patients, respectively, and there was no significant difference among diseases. Real-time quantitative PCR showed no significant difference in the relative concentrations of pks-positive bacterial DNA among diseases. Age, gender, location of CRC, CRC staging, or k-ras gene status was not associated with pks prevalence.
Conclusions
Although the method of collecting fecal DNA from colonic lavage samples was safe and technically feasible, factors other than pks-positive bacteria appear to play more important roles in CRC development in this cohort.
Similar content being viewed by others
Background
Infectious microorganisms are often thought to be associated with human carcinogenesis. For example, Helicobacter pylori, a Gram-negative bacterium chronically residing in the human stomach, is referred to as a definite carcinogen of gastric cancer [1,2,3]. Human hepatitis B and C viruses are also established as causative agents of hepatocellular carcinoma [4, 5]. Similar association is also established between some human papilloma viruses and carcinoma of the cervix uteri [6].
Colorectal cancer (CRC) is the fourth leading cause of cancer-related deaths in the world [7], and its prevention and early detection are among the most urgent needs regarding public health. Human colon is colonized by more than 1014 bacteria which can be classified into at least 1000 species, making up what is called the microbiota, and it would be natural to assume that some of these bacteria are associated with the pathogenesis of human CRC. Recently, several studies have indicated that certain strains of Escherichia coli (E. coli) possessing a gene cluster named the pks island might have a causative role in human CRC development [8, 9]. E. coli, a Gram-negative, facultative anaerobic rod, a member of the family Enterobacteriaceae, is found widely in the gastrointestinal tract of many mammals, including almost all humans. This species is further divided into phylogenetic groups A, B1, B2, and D. Most strains belonging to group B2 are known for their extraintestinal pathogenic nature causing urinary tract infection, sepsis, and newborn meningitis in humans, which are called extraintestinal pathogenic E. coli (ExPEC), whereas strains in groups A and D are mostly nonpathogenic commensals or pathogens which mainly cause intestinal disorders presenting as diarrhea [10, 11]. The pks island, made up of approximately 54,000 base pairs, consists of genes coding three nonribosomal peptide megasynthases (NRPS), three polyketide megasynthases (PKS), and two hybrid NRPS/PKS megasynthases [12], and is thought to produce a peptide–polyketide genotoxin named colibactin. This pathogenic island is found mostly in phylogenetic group B2 E. coli [9, 13, 14], although there are also a much smaller number of pks-positive strains in group B1 E. coli and other bacterial species in the family Enterobacteriaceae [15].
Previous in vitro experiments showed that pks-positive E. coli induces DNA double-strand breaks and transient G2-M cell cycle arrest in its host mammalian cells, whereas pks-negative E. coli does not [13, 16]. Those infected host cells can survive after incomplete DNA repair, resulting in higher mutation rates, presumably leading to tumorigenesis. They also showed that pks-positive E. coli loses this genotoxic property by knocking out the pks island, while pks-negative E. coli acquires genotoxicity by introducing a gene construct containing the pks island [13, 16]. In vivo studies used colitis-susceptible interleukin-10-deficient mice under germ-free condition, and when they were associated with either pks-positive E. coli or pks-negative E. coli, together with azoxymethane, the group of mice receiving pks-positive E. coli showed significantly greater increase in colon tumor occurrence and invasiveness compared to the other group, while the severity of colitis was similar between the two groups [8]. When host cells are infected with greater number of pks-positive E. coli, they show senescence-associated secretory phenotype, by which the cells secrete growth factors such as hepatocyte growth factor, stimulating non-infected neighboring cells to proliferate, also possibly causing tumor formation [17].
Concerning humans, two studies from Europe showed that pks-positive E. coli are more prevalent in colon tissue specimens derived from CRC patients compared to those from non-CRC controls. Arthur et al. studied on tissue specimens from 21 CRC patients and 24 controls and found pks-positive E. coli from 67 and 21% of each category, respectively [8]. Likewise, Buc et al. studied on tissue specimens from 38 CRC patients and 31 diverticulosis patients and found pks-positive E. coli from 55 and 19%, respectively [9]. Thus, the pks island may act as a tumor promoter for CRC, and could be used as a predictive biomarker for CRC development.
In the present study, we sought to clarify the difference in pks prevalence between CRC patients and non-CRC controls in the Japanese population, by using non-invasive endoscopic sample collection technique.
Methods
Patients
Patients who received total colonoscopy at the University of Tokyo Hospital from January 2014 through May 2015 were asked for participation, and written consent was obtained from every patient who agreed to participate. Clinical characteristics and history of previous and present endoscopic findings of each patient were recorded. Patients were classified into three disease categories: CRC, adenoma, and control; regardless of whether the disease was diagnosed by previous or present session of colonoscopy. In all CRC patients, endoscopic diagnosis was confirmed by pathological examination of previous or concomitant biopsy samples. k-ras gene status of the tumor was determined by direct sequencing of tumor DNA extracted from paraffin-embedded tumor tissues.
This study was approved by the ethics committee of the University of Tokyo (approval number 10329).
DNA extraction from colonic lavage samples and biopsy samples
Ten milliliters of residual suspension in the colonic lumen after lavage was collected during colonoscopy of every patient, using a syringe connected to the endoscope. The samples were then transiently stored and transported at 4 °C. Solid component within each suspension sample, which contained bacterial cells, was isolated by centrifugation and was washed with phosphate buffer saline for three times, in order to remove possible water-soluble distractors such as polyethylene glycol contained in oral bowel preparation solution. Then total bacterial DNA was extracted using QIAamp DNA Stool Mini Kit (QIAGEN), and was stored at −30 °C until use.
Biopsy samples were washed with phosphate buffer saline in order to remove mucus and mucus-associated bacteria. DNA was extracted from the tissue and co-existing mucosa-associated bacteria using QIAamp DNA Mini Kit (QIAGEN), and was stored at −30 °C until use.
PCR analysis
Both conventional qualitative PCR and real-time quantitative PCR were used to evaluate fecal DNA samples. In order to detect pks-positive bacterial DNA, primers were used to amplify a 283 bp sequence in clbB gene of E. coli, a part of the pks island, with the forward primer 5′-GCGCATCCTCAAGAGTAAATA-3′ and the reverse primer 5′-GCGCTCTATGCTCATCAACC-3′ [8]. Primers to amplify a 147 bp sequence in E. coli-specific gene uidA with the forward primer 5′-TGGTAATTACCGACGAAAACGGC-3′ and the reverse primer 5′-ACGCGTGGTTACAGTCTTGCG-3′, and those to amplify a 144 bp universal bacterial sequence in 16S rRNA gene with the forward primer 5′-GGTGAATACGTTCCCGG-3′ and the reverse primer 5′-TACGGCTACCTTGTTACGACTT-3′, were also used in order to detect total E. coli DNA and total bacterial DNA, respectively [18, 19].
Twenty microliters of reaction mixture for conventional qualitative PCR consisted of 1 μL of template DNA solution, each 1 μL of 10 μmol/L forward and reverse primer solutions, deoxynucleotide mixture, the DNA polymerase AmpliTaq Gold (Applied Biosystems), and buffer according to the manufacturer’s instructions. The PCR conditions were 7 min at 95 °C, 35 cycles of 30 s at 95 °C, 30 s at 55 °C, and 30 s at 72 °C, and 7 min at 72 °C.
Patients were considered pks-positive when their fecal bacterial DNA tested pks-positive after conventional qualitative PCR, and for each disease category, pks prevalence was defined as the proportion of the number of pks-positive patients to that of total patients in that category.
We have isolated several E. coli strains from patients. One of these isolates was positive for clbB gene, uidA gene, and 16S rRNA gene by conventional qualitative PCR and DNA sequencing. This isolate was also positive for clbQ and clbA genes, which are located near the 5′ and 3′ terminals of the pks island, respectively, thus suggesting this isolate contained the entire sequence of the pks island [14]. Primers to amplify a 308 bp sequence in clbQ gene with the forward primer 5′-GCACGATCGGACAGGTTAAT-3′ and the reverse primer 5′-TAGTCTCGGAGGGATCATGG-3′, and those to amplify a 342 bp sequence in clbA gene with the forward primer 5′-AAGCCGTATCCTGCTCAAAA-3′ and the reverse primer 5′-GCTTCTTTGAGCGTCCACAT-3′, were used. We used this E. coli isolate as the positive control for the pks island. As the negative control for the pks island and the positive control for uidA gene and 16S rRNA gene, str. K-12 substr. MG1655 was used.
Twenty microliters of reaction mixture for real-time quantitative PCR consisted of 2 μL of template DNA solution, each 0.6 μL of 10 μmol/L forward and reverse primer solutions, and the FastStart Universal SYBR Green Master (ROX) mix (Roche) which contains the DNA polymerase. The PCR conditions were 10 min at 95 °C, 40 cycles of 15 s at 95 °C and 60 s at 60 °C, followed by an appropriate dissociation phase.
For each fecal bacterial DNA sample, relative concentration of pks-positive bacterial DNA was defined as the proportion of calculated concentration of pks-positive DNA given by real-time quantitative PCR to that of total bacterial DNA.
Statistical analysis
Continuous variables were compared using the Mann–Whitney U test, and categorical variables were compared using the χ2 test. All tests were two-sided, and a p value <0.05 was considered significant.
Results
Detection threshold of pks-positive bacterial DNA
First we aimed to validate the quality of DNA detection system from colonic lavage samples by using qualitative PCR. We mixed pks-positive and -negative E. coli DNA together at various ratios, keeping the sum concentration at 500 pg per PCR sample. On conventional qualitative PCR, samples containing 0.1 pg or more of pks-positive control DNA tested positive (Fig. 1). Given that E. coli genome weighs approximately 5 × 10−15 g per copy, this result means that a PCR sample containing DNA from 20 or more pks-positive E. coli, or a colonic lavage sample (10 mL) containing 1 × 104 or more of them gives a positive result in conventional qualitative PCR.
Conventional qualitative PCR to detect various amounts of pks-positive control E. coli DNA using primers for clbB (top) and uidA (bottom) genes. N negative control for clbB gene (derived from str. K-12 substr. MG1655), P positive control for clbB gene (referred to in the “Methods” section); numerals, amount (pg/sample) of pks-positive E. coli DNA. Samples containing 0.1 pg or more of pks-positive control DNA tested positive
Correlation between clbB gene and the whole pks Island
In order to use clbB gene as a surrogate marker of the whole pks island, we examined clbQ and clbA genes, which are located very near to the 5′ and 3′ terminals of the pks island, respectively. We selected nine clbB-positive DNA samples derived from nine different E. coli strains, and found that all of them were also positive for both clbQ and clbA genes (Fig. 2), which was consistent to a previous study [14]. Therefore, in this study, we used clbB gene as a surrogate marker of the whole pks island.
Conventional qualitative PCR to detect clbB, clbQ, and clbA genes from nine DNA samples derived from nine different E. coli strains. N negative control for clbB gene (derived from str. K-12 substr. MG1655), P positive control for clbB gene (referred to in the “Methods” section)
Correlation between colonic lavage samples and biopsy samples
Previous studies aimed to discuss the prevalence of pks-positive E. coli among mucosa-associated bacteria by using surgical specimens or biopsy samples of the colon. In order to ensure that this study using colonic lavage samples can be compared with those previous studies, we examined the correlation between the pks status of the colonic lavage sample and the biopsy sample of the same patients. We obtained a colonic lavage sample and a biopsy sample concurrently during one session of colonoscopy per patient, and collected 13 pairs of colonic lavage samples and biopsy samples from 13 patients. Of those, 12 patients (92%) showed the same pks status by colonic lavage sample and biopsy sample (3 patients were both pks-positive and the other 9 were both pks-negative). One patient provided a pks-positive colonic lavage sample and a pks-negative biopsy sample (Table 1). Correlation between the pks status by the two sampling methods was good, with the κ coefficient 0.806. This result suggested the usefulness of colonic lavage samples as a surrogate for biopsy samples—as far as pks status is concerned.
Detection and quantification of the pks-positive E. coli by colonic lavage samples
Colonic lavage samples were collected during colonoscopy from 98 patients including 35 from CRC patients, 37 from adenoma patients, and 26 from controls. Each disease category contains past history of respective disease. Gender, age, and amount of total bacterial DNA extracted from each colonic lavage samples are listed in Table 2. Male prevalence was significantly greater among the CRC patients compared to the controls, but otherwise there was no significant difference.
The pks prevalence in each disease category was determined by conventional qualitative PCR. It was 43 and 51% in CRC and adenoma patients, respectively. Neither of them showed significant difference from that in controls, which was 46% (the p value was 0.798 and 0.685, respectively) (Table 2). We also calculated pks prevalence according to their present endoscopic findings, and no difference was observed either (data not shown).
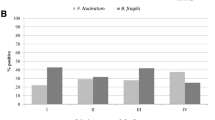
Relative concentration of pks-positive bacterial DNA in each fecal sample was determined by real-time quantitative PCR. The results are shown in Fig. 3, according to disease category. It ranged from 0 to 0.097 (median 0.00019) in CRC patients, and from 0 to 0.24 (median 0.00025) in adenoma patients. Neither of them showed significant difference from that in controls, which ranged from 0 to 0.74 (median 0.00014) (the p value was 0.836 and 0.570, respectively). We also determined relative pks concentration according to present endoscopic findings, and there was no significant difference either (data not shown).
There was no correlation between patient age and relative pks concentration (Fig. 4a). Relative pks concentration ranged from 0 to 0.74 (median 0.00018) in male patients and ranged from 0 to 0.16 (median 0.00026) in female patients, which produced no significant difference (Fig. 4b).
Post-hoc subgroup analysis (CRC patients)
We performed post hoc subgroup analysis on pks prevalence in CRC patients for whom some additional information on their CRC was available (Table 3). pks prevalence was not significantly different between patients with right-sided CRC (located in cecum, ascending colon, or transverse colon) and those with left-sided CRC (located in descending colon, sigmoid colon, or rectum). We also compared patients whose tumor invading only the mucosa (M) or submucosa (SM), and those with tumor invading the muscularis propria (MP) or further, but no significant difference in pks prevalence was detected. Half of the patients with k-ras mutant CRC were pks-positive, while none of those with k-ras wild-type CRC were pks-positive, although this difference in pks prevalence did not reach to statistical significance. The presence of previous or ongoing treatment against CRC, or the presence of colonic obstruction due to tumor did not significantly affect pks prevalence.
Discussion
Our present study is the first to investigate the difference in pks prevalence between CRC patients and non-CRC controls in the Japanese population. It is also unique in that it used colonic lavage samples derived during colonoscopy, and the samples were collected and stored in a uniform method performed by a limited number of investigators with good understanding of the study, which might be advantageous compared to asking patients to collect their stool for themselves. Using colonic lavage samples is less invasive compared to biopsy, which also makes this method easy to practice in clinical settings.
This study showed that the prevalence of pks-positive E. coli was not significantly higher in CRC patients compared to controls. Based on the findings in the previous study [8], we had assumed that pks prevalence in CRC and control patients would be 67 and 21%, respectively. According to this assumption and a two-sided α level of 0.05, 35 CRC patients and 26 controls in the present study give the statistical power of 92%, which makes it likely that the result in this study is a true negative.
Using colonic lavage samples, pks prevalence in CRC patients was 43%. This value is lower compared to those of CRC patients in two previous studies from Europe using colon tissue specimens, which are 67 and 55%, respectively (Table 4) [8, 9]. On the other hand, pks prevalence in our non-CRC controls (46%) appears higher than their counterparts in those studies (21 and 19%, respectively). This discrepancy in pks prevalence might simply reflect the difference between mucosa-associated bacteria and those floating in the gut lumen, but in two studies from the United States using stool instead of tissue specimens showed pks prevalence of 32 and 20% [14, 20], which are not very different from those using tissue specimens. We also evaluated pks prevalence using colonic lavage samples and concurrent biopsy samples from 13 patients, which indicated good correlation. Another possible explanation to this discrepancy is the microbial alteration by colonic lavage procedure before colonoscopy. However, several previous reports show such changes rarely overcome interpersonal differences in bacterial profile at phylum or genus level [21, 22].
We should also note that healthy Japanese population seems to show relatively high prevalence of phylogenetic group B2 strains among their stool E. coli compared to their Western counterparts. In one report from Tokyo, Japan, group B2 strain accounted for 44% of all E. coli isolates [23]. On the other hand, this value was 16, 32, 33, and 37% in one report from France [24], 29% in another from the same country [24], and 48% in one from the United States [25]. Taking into account that most pks-positive bacteria belong to phylogenetic group B2 E. coli, this can lead to high prevalence of pks-positive bacteria among healthy Japanese people, including non-CRC controls in this study.
Post-hoc subgroup analysis on CRC patients in this study showed no significant association between clinical parameters and pks prevalence. However, it might be worth noting that pks-positive E. coli was detected only in k-ras mutant CRC patients, although the difference did not reach to statistical significance due to small sample size. It might imply a possibility that pks-positive E. coli can induce k-ras gene mutation through its genotoxic property.
Conclusions
Although the method of collecting fecal DNA from colonic lavage samples was safe and technically feasible, factors other than pks-positive bacteria appear to play more important roles in CRC development in the Japanese population.
References
Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–9.
De Vries AC, Van Grieken NC, Looman CW, Casparie MK, de Vries E, Meijer GA, Kuipers EJ. Gastric cancer risk in patients with premalignant gastric lesions: a nationwide cohort study in the Netherlands. Gastroenterology. 2008;134:945–52.
Sakitani K, Hirata Y, Watabe H, Yamada A, Sugimoto T, Yamaji Y, Yoshida H, Maeda S, Omata M, Koike K. Gastric cancer risk according to the distribution of intestinal metaplasia and neutrophil infiltration. J Gastroenterol Hepatol. 2011;26:1570–5.
Fattovich G, Bortolotti F, Donato F. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol. 2008;48:335–52.
Morgan RL, Baack B, Smith BD, Yartel A, Pitasi M, Falck-Ytter Y. Eradication of hepatitis C virus infection and the development of hepatocellular carcinoma: a meta-analysis of observational studies. Ann Intern Med. 2013;158:329–37.
Nobbenhuis MA, Walboomers JM, Helmerhorst TJ, Rozendaal L, Remmink AJ, Risse EK, van der Linden HC, Voorhorst FJ, Kenemans P, Meijer CJ. Relation of human papilloma virus status to cervical lesions and consequences for cervical-cancer screening: a prospective study. Lancet. 1999;354:20–5.
Stewart BW, Wild CP. World Cancer Report 2014. IARC. 2014.
Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan T-J, Campbell BJ, Abujamel T, Dogan B, Rogers AB. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–3.
Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, Pezet D, Bonnet R. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE. 2013;8:e56964.
Ochman H, Selander RK. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–3.
Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventre A, Elion J, Picard B, Denamur E. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology. 2001;147:1671–6.
Bian X, Plaza A, Zhang Y, Muller R. Two more pieces of the colibactin genotoxin puzzle from Escherichia coli show incorporation of an unusual 1-aminocyclopropanecarboxylic acid moiety. Chem Sci. 2015;6:3154–60.
Nougayrède J-P, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–51.
Johnson JR, Johnston B, Kuskowski MA, Nougayrede JP, Oswald E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J Clin Microbiol. 2008;46:3906–11.
Putze J, Hennequin C, Nougayrede JP, Zhang W, Homburg S, Karch H, Bringer MA, Fayolle C, Carniel E, Rabsch W, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun. 2009;77:4696–703.
Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. 2010;107:11537–42.
Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L, Sauvanet P, Darcha C, Dechelotte P, Bonnet M, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63:1932–42.
Tantawiwat S, Tansuphasiri U, Wongwit W, Wongchotigul V, Kitayaporn D. Development of multiplex PCR for the detection of total coliform bacteria for Escherichia coli and Clostridium perfringens in drinking water. Southeast Asian J Trop Med Public Health. 2005;36:162–9.
Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41.
Gomez-Moreno R, Robledo IE, Baerga-Ortiz A. Direct detection and quantification of bacterial genes associated with inflammation in DNA Isolated from stool. Adv Microbiol. 2014;4:1065–75.
Jalanka J, Salonen A, Salojarvi J, Ritari J, Immonen O, Marciani L, Gowland P, Hoad C, Garsed K, Lam C, et al. Effects of bowel cleansing on the intestinal microbiota. Gut. 2015;64:1562–8.
Lavelle A, Lennon G, O’Sullivan O, Docherty N, Balfe A, Maguire A, Mulcahy HE, Doherty G, O’Donoghue D, Hyland J, et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut. 2015;64:1553–61.
Obata-Yasuoka M, Ba-Thein W, Tsukamoto T, Yoshikawa H, Hayashi H. Vaginal Escherichia coli share common virulence factor profiles, serotypes and phylogeny with other extraintestinal E. coli. Microbiology. 2002;148:2745–52.
Escobar-Paramo P, Grenet K, Le Menac’h A, Rode L, Salgado E, Amorin C, Gouriou S, Picard B, Rahimy MC, Andremont A, et al. Large-scale population structure of human commensal Escherichia coli isolates. Appl Environ Microbiol. 2004;70:5698–700.
Zhang L, Foxman B, Marrs C. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J Clin Microbiol. 2002;40:3951–5.
Authors’ contributions
TS collected, analyzed, and interpreted the patient data regarding colonic lavage samples, biopsy samples, and endoscopic findings, and was a major contributor in writing the manuscript. All authors participated in collecting patient data. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Ethical approval and consent to participate
This study was approved by the ethics committee of the University of Tokyo (Approval Number 10329). Patients were asked for participation, and written consent was obtained from every patient who agreed to participate.
Funding
This study was supported by JSPS KAKENHI Grant Numbers 26460962 to SY, and 17K09347 to TS.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Shimpoh, T., Hirata, Y., Ihara, S. et al. Prevalence of pks-positive Escherichia coli in Japanese patients with or without colorectal cancer. Gut Pathog 9, 35 (2017). https://doi.org/10.1186/s13099-017-0185-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13099-017-0185-x