Abstract
Background
Dengue, chikungunya and Zika viruses (DENV, CHIKV and ZIKV) are transmitted in sylvatic transmission cycles between non-human primates and forest (sylvan) mosquitoes in Africa and Asia. It remains unclear if sylvatic cycles exist or could establish themselves elsewhere and contribute to the epidemiology of these diseases. The Caribbean island of St. Kitts has a large African green monkey (AGM) (Chlorocebus aethiops sabaeus) population and is therefore ideally suited to investigate sylvatic cycles.
Methods
We tested 858 AGM sera by ELISA and PRNT for virus-specific antibodies and collected and identified 9704 potential arbovirus vector mosquitoes. Mosquitoes were homogenized in 513 pools for testing by viral isolation in cell culture and by multiplex RT-qPCR after RNA extraction to detect the presence of DENV, CHIKV and ZIKVs. DNA was extracted from 122 visibly blood-fed individual mosquitoes and a polymorphic region of the hydroxymethylbilane synthase gene (HMBS) was amplified by PCR to determine if mosquitoes had fed on AGMs or humans.
Results
All of the AGMs were negative for DENV, CHIKV or ZIKV antibodies. However, one AGM did have evidence of an undifferentiated Flavivirus infection. Similarly, DENV, CHIKV and ZIKV were not detected in any of the mosquito pools by PCR or culture. AGMs were not the source of any of the mosquito blood meals.
Conclusion
Sylvatic cycles involving AGMs and DENV, CHIKV and ZIKV do not currently exist on St. Kitts.

Similar content being viewed by others
Background
Chikungunya, dengue and Zika are arboviral diseases that are transmitted by the anthrophilic mosquitoes, Aedes aegypti and/or Aedes albopictus in an urban transmission cycle resulting in epidemics and pandemics in tropical and subtropical regions of the world [1,2,3]. The aetiological chikungunya (CHIKV), dengue (DENV) and Zika viruses (ZIKV) evolved in non-human primates (NHPs) and sylvatic mosquitoes in the forests of Africa in the case of CHIKV and ZIKV, and Asia in the case of DENV [4,5,6]. In the forests, primatophilic forest mosquitoes maintain the viruses in sylvatic (NHP-mosquito-NHP) transmission cycles which continue to this day [3, 7,8,9]. It remains an outstanding question whether sylvatic cycles of these arboviruses are present elsewhere in the world where there are similar non-human primate vertebrate hosts and mosquitoes [8, 10,11,12]. Furthermore, some researchers have identified tropical islands as ‘hotspots’ for arboviral emergence [13].
On the Caribbean island of St. Kitts there is a large population of wild and captive African green monkeys (AGMs) (Chlorocebus aethiops sabeus). People on the island were affected by the chikungunya pandemic in 2014 and the Zika pandemic in 2016 [14, 15]. Dengue is hyperendemic in the region and there are periodic outbreaks on St. Kitts, most recently in 2008 [16]. While there are no accurate seroprevalence data on arboviral infections of people on St. Kitts, studies from nearby islands show very high CHIKV exposure rates of 16.9% [17] on St. Maarten and 25% in Puerto Rico [18]. Similar exposure rates have been suggested for ZIKV [19]. The confirmed presence of arboviral disease in people, a suitable vertebrate host (AGMs) and a diverse mosquito community [20] suggests there is potential for sylvatic transmission on St. Kitts.
To investigate this possibility, we used previously established techniques [7, 21,22,23,24] involving serology to determine exposure of the island’s AGMs to arboviruses, and RT-qPCR and viral isolation in cell culture to determine the presence of arboviruses in potential vector mosquito genera. Additionally, DNA extracted from blood meals of engorged female mosquitoes was analyzed by PCR and sequenced to determine which mosquitoes had fed on AGMs and/or people.
Methods
Study area
St. Kitts is a 168 km2, geographically isolated, volcanic, Caribbean island located in the Lesser Antilles (17.33°N, 62.75°W). It has a population of approximately 40,000 people mostly inhabiting urban Basseterre, the capital, and a string of small village communities distributed along the main coastal road that circles the island. Rainforest covers the uninhabited, steep volcanic slopes in the center of the island, surrounded by lower gentler slopes consisting mostly of abandoned sugar cane fields or arable farmlands. The south east of the island is primarily an arid peninsula covered mainly in scrub with beaches, mangroves, and salt-ponds. AGMs were introduced to the island in the 1700s during the slave trade and are abundant with an estimated population of 55,000 [25,26,27]. Due to changes in land use and their adaptable and opportunistic nature they are now widely distributed, commonly encountered, a tourist attraction and also a problem for local farmers whose crops suffer from their destructive behaviour [25]. They are semi-arboreal, roosting in trees at night, but highly mobile on the ground during the day when they forage through many of the ecosystems on the island [28]. While foraging for food and water, which can be over large areas depending on availability that varies with season, it would be anticipated that AGMs should come into contact with, and potentially host a wide variety of mosquitoes although we know of no data on the species involved.
Determining the infection status of the AGMs
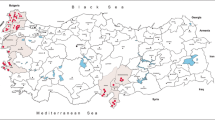
Between January 2013 and March 2019, we obtained 851 convenience samples of sera from AGMs undergoing routine health screening after being trapped using accepted procedures [26, 27] for two primate research facilities on St. Kitts. We also obtained seven sera from AGMs trapped and immediately released by professional monkey trappers for our study (Additional file 1: Text S1). Monkeys were captured across five different land covers that we identified on the island: agricultural; mangrove; urban; rainforest; and scrub (Fig. 1 and Table 1) [20].
Approximate capture locations by parish and land cover of AGMs on St. Kitts. Numbers of AGMs captured in each land cover indicated by a coloured pie chart. Key: brown, agricultural; light blue, mangrove; light green, rainforest; dark yellow, scrub; light grey, urban. Parish boundaries are indicated by a grey line
Sera from 268 of the AGMs captured prior to the Zika outbreak in 2016 were screened for IgG to DENV and CHIKV with commercial ELISA kits (Panbio Dengue IgG Indirect Elisa. Standard Diagnostics Inc., Yongin-si, Republic of Korea; Anti-Chikungunya virus ELISA (IgG). Euroimmune AG, Lübeck, Germany) following the manufacturer’s instructions. They were also tested for IgG to DENV and CHIKV at the National Reference Centre for Arboviruses in Marseille, France (IL-G) with an in-house ELISA for with peroxidase labelled anti-monkey IgG (KPL, Gaithersburg, MD, USA) as a secondary antibody. The remaining 590 sera were also screened for antibodies to DENV and CHIKV with the commercial ELISA kits described above and additionally for ZIKV IgG antibodies (Monkey Zika Virus IgG (ZV-IgG) ELISA Kit, MyBioSource, Inc., San Diego, CA, USA). Sera with positive or equivocal ELISA results were repeated and then tested by Plaque Reduction Neutralization Test (PRNT) [29,30,31] on Vero cells (ATCC #CCL-81) with a cutoff value of 90% (PRNT90). Neutralization curves were generated using GraphPad Prism software, and the resulting data were analyzed by nonlinear regression to estimate the dilution of serum required to inhibit 90% of infection. We considered an animal to have a confirmed ZIKV exposure if ZIKV PRNT90 was at least 20 and a ratio of ZIKV PRNT90 to DENV PRNT90 titre of at least 4.
Determining the infection status of potential arboviral vector mosquito species
Mosquito collections
From September 2017 to March 2019 we conducted a comprehensive mosquito survey [20] and captured mosquitoes each month for a 48-h period (excluding December 2017, April 2018, and December 2018) in all five land covers described above using carbon dioxide (CO2) baited CDC light traps (J.W. Hock, Gainesville, FL, USA) and/or Biogents Sentinel 2 traps (BGS) (Biogents AG, Regensburg, Germany) to capture a broad range of mosquito species and target host-seeking mosquitoes which are most likely to have fed on mammals [20]. Since it was not possible for us to trap mosquitoes in the often inaccessible areas where AGMs sleep and forage, which both vary considerably, we used sites described in the mosquito survey [20] and these were set within 100–200 m of known AGM trapping locations where possible. Trapped mosquitoes were transported to the research laboratory of Ross University School of Veterinary Medicine (RUSVM), Basseterre, St. Kitts, and stored at − 80 °C for later identification. Potential arbovirus vector mosquito species (Aedes aegypti, Aedes taeniorhynchus, Culex quinquefasciatus and unidentified Aedes and Culex spp.) were identified using standard morphological keys [32,33,34]. For arboviral testing (below), 1–50 individuals of each species were pooled according to location where they were trapped, month, and sex.
Mosquito processing
Mosquito pools were homogenized in a 2 ml microcentrifuge tube containing 3–4, sterile, steel ball bearings (4 mm in diameter) with 600 µl of minimum essential media (MEM, Gibco, Waltham, MA, USA) with 1% penicillin and streptomycin (Penicillin-Streptomycin, 10,000 U/ml, Gibco Waltham, MA, USA) and agitated for 5 min using a vortex. Homogenates were clarified by centrifugation (5 min at 14,000× rpm) at 4 °C and the supernatant, approximately 300–500 µl, filtered (0.22 µm syringe filter Millipore Millex™ Sterile Syringe Filters. Merck KGaA, Darmstadt, Germany) and stored at − 80 °C.
RNA extraction and RT-qPCR
After thawing at room temperature, RNA was extracted from 100 µl of the mosquito lysate using the RNEasy Mini Kit (Qiagen, Hilden, Germany) and analyzed by RT-qPCR using the ZDC (Zika, Dengue and Chikungunya) Multiplex RT-PCR assay (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions on an ABI 7500 Fast Dx Real-Time PCR system (Applied Biosystems, Hercules, CA, USA).
Cell culture for viral isolation
Mosquito lysates (100 µl) were inoculated into 24 well cell culture plates seeded with approximately 5 × 104 Vero cells in 0.5 ml of MEM with 1% Fetal Bovine Serum (Gibco), 1% Penicillin-Streptomycin (Gibco), 1% glutamine (Gibco) and 0.1 % Amphotericin B (Gibco). Following gentle agitation on a rocker for 1 h to allow virus adsorption, 1ml of MEM (as above) was added to each well and the plates incubated at 37 °C for 7–10 days. Every day the cells in each well were monitored for cytopathic effects with an inverted microscope (CKX53, Olympus, Tokyo, Japan).
Blood-meal analysis
Engorged blood-fed female mosquitoes (n = 122) were retained individually and their abdomens aseptically separated from the head and thorax by sharp dissection. Their DNA was extracted using a DNEasy Blood Mini Kit (Qiagen, Hilden, Germany) and used in a qPCR with primers designed to anneal to the hydroxymethylbilane synthase (HMBS) gene as described by Wei et al. [35]. DNA extracted from the whole blood of five AGMs were used as positive controls and their sequences (ELIM BIOPHARM, Hayward, CA, USA) aligned with Clustal Omega [36] to obtain a 222 nucleotide sequence for the HMBS gene of Chlorocebus aethiops sabeus (Additional file 2: Text S2) (BankIt2363830 AGM_seq MT742560). Amplicons obtained from the blood-fed mosquitoes were also sequenced and raw sequence data was compared with the AGM HMBS (BankIt2363830 AGM_seq MT742560) and human HMBS sequences NG_008093 on GenBank using Clustal Omega.
Results
Serology of AGMs
The 268 sera tested with an in-house ELISA in France and with commercial ELISA test kits for antibodies to DENV and CHIKV all gave negative results. The remaining 590 samples tested with the commercial kits for antibodies to DENV, CHIKV and ZIKV were all negative except for 10 (1.7%) that were positive for ZIKV IgG. On subsequent confirmatory testing by PRNT all ten tested negative for ZIKV and DENV neutralizing antibodies using PRNT90.
Arbovirus detection
We captured 9704 individual mosquitoes representing 10 of the 14 mosquito species across all 6 genera previously documented on the island [20, 37] (Additional file 3: Table S1). Approximately half (3000–4000 individuals, 190 pools) of the mosquitoes from non-urban land covers were trapped within 100–200 m of where AGMs were blood-sampled for serology. All the 513 pools of mosquitoes (Tables 2, 3) tested by RT-qPCR for CHIKV, DENV and ZIKV were negative. Furthermore, all virus isolation attempts in Vero cell cultures were negative.
Blood-meal analysis
Of the 122 blood meals we tested with the HMBS PCR, we obtained useable sequence data from 106 sequences (including 13 from Ae. aegypti, the only known vector of DENV, CHIKV and ZIKV recorded on St. Kitts) that aligned in the polymorphic 286 base pair (bp) target region of the AGM consensus sequence and human HMBS reference gene (NG_008093.1) [31] containing 5 reliable single nucleotide polymorphisms (SNP) and 2 deletions (Additional file 2: Text S2). Of these, one blood meal from Ae. taeniorhynchus caught in scrub land cover (mosquito 106) had 100% sequence match (227 bp) with the human reference HMBS gene (NG_008093.1) compared with a 95.5% (213 bp) with the AGM HMBS gene sequence we produced (BankIt2363830 AGM_seq MT742560). Similarly, we also considered 3 more blood meals with lower sequence matches (83.25–97.64%) from a Culex spp. in the mangrove (mosquito 43) and two unidentifiable urban mosquitoes (mosquitoes 64 and 65) to match best with the human reference gene (Additional file 2: Text S2).
Discussion
Our data do not provide robust evidence for sylvatic transmission of CHIKV, DENV or ZIKV between AGMs on St. Kitts. By capturing and testing both AGMs and sympatric mosquitoes for evidence of arboviral infections we emulated studies from Africa and Asia [7, 21,22,23,24] that have produced reliable (and some of the original) data on the existence of sylvatic transmission cycles of these arboviruses [4]. It is of note, however, that some studies on sylvatic cycles relied solely on antibody detection in NHPs [11, 38,39,40,41,42] to provide evidence of arboviral exposure. In other cases, evidence for these cycles has only been based on the presence of arboviruses in mosquitoes captured from sylvatic habitats [3, 7, 21, 43, 44].
The actual population of AGMs on St. Kitts is unknown, but estimates are up to 55,000 [25, 26]. Calculations show that detecting a 1% disease prevalence in this highest population estimate of 55,000 animals at a 95% confidence level requires a sample size of 332 (assuming 100% test sensitivity and specificity) [45, 46]. Our convenience sample of 858 monkey sera would then appear to have been more than adequate to detect low seroprevalences.
Our inability to detect significant levels of antibodies to CHIKV, DENV, and ZIKV provides strong evidence that the AGMs on St. Kitts are not, or only very infrequently, exposed to these viruses. We contemporaneously sampled AGMs during chikungunya (2014) and Zika (2016) epidemics and it would seem reasonable to assume this would have given us a good opportunity to detect AGM infections if there was spillover.
Although the PRNT remains the gold standard serological test for the diagnosis of infections with the different arboviruses, it is time consuming, requires highly trained staff, live viruses, reliable controls and a BSL-2 laboratory (BSL-3 in the case of CHIKV). The IgG ELISAs we used for screening produce reliable negative results and are frequently used in experimental arboviral vaccine models involving NHPs to prove freedom from exposure [47,48,49,50]. They have proved to be reproducible and sensitive [51] in arboviral studies involving various NHPs in Africa, Asia and the Americas [38,39,40,41,42, 52,53,54]. The ELISAs are well suited for large-scale screening for previous infections in field studies because wild infected NHPs mount a robust antibody response although only viraemic for 1–7 days and show no obvious clinical signs [4]. The IgG antibodies can be detected for years post-infection and the commercial kits for humans are widely available and can be adapted for use in NHPs [4]. Our findings confirm the reliability of ELISA testing in the general screening of a population with all of the randomly selected 268 ELISA negative sera also being negative when tested by the National Reference Centre for Arboviruses in Marseille, France. The problem with ELISAs is a lack of specificity because of cross reactivity of antibodies against closely related viruses, although this can also be an advantage as it increases the kits’ screening potential [55]. Sera from the 10 AGMs that were ZIKV IgG antibody-positive by ELISA were confirmed ZIKV negative by PRNT90. This maybe within the limits of the test specificity or because of exposure to another undifferentiated Flavivirus. We did not have access to the variety of other flaviviruses that occur in the region for more definitive PRNT testing of this sample. The other flaviviruses that have been described in the Caribbean region and have high potential for cross reactivity include West Nile virus (WNV), and Spondweni virus (SPONV). WNV has been isolated from humans, birds and mosquitoes (suggesting active transmission) in Puerto Rico in 2007 [56], and recently 10.7% of equids on St. Kitts were reported to be seropositive to WNV [57]. SPONV is difficult to distinguish clinically and serologically from ZIKV infection and had only been recorded in Africa until 2016 when it was isolated from Cx. quinquefasciatus from Haiti [58]. With respect to alphaviruses, Mayaro virus (MAYV) can serologically cross react with CHIKV and was first discovered in Trinidad in 1954 and more recently antibodies have been detected in NHPs in Panama and French Guiana [4]. Further work is underway in our laboratories to identify other arboviruses such as these that might be circulating on St. Kitts.
Although our trapping methods would have influenced the numbers and diversity of mosquitoes we caught, in testing the mosquitoes our trapping selected for, including mosquitoes from areas where the monkeys we serosurveyed were trapped, we found no evidence for the presence of CHIKV, DENV or ZIKV. We used RT-qPCR and virus isolation in cell culture to improve detection rates as has been done, either singly or in combination, to detect arboviruses in mosquitoes in large studies in Africa [59, 60]. The RT-qPCR is very specific and sensitive, detecting as little as one infected mosquito in a pool of 5000, and enables high throughput rapid results with less reliance on a cold chain [60]. Virus isolation is less sensitive than RT-qPCR, detecting viable virus which requires the presence of a robust cold chain, specialized laboratory facilities, skilled labour and time, at least a week.
Our inability to demonstrate arboviruses in the mosquitoes was not unexpected in light of the lack of seropositivity in the AGMs and, in the case of the urban mosquitoes we tested, the fact that only low numbers of dengue, chikungunya and Zika cases have been reported in people on St. Kitts. Low viral prevalence in mosquitoes makes arboviral detection difficult even during disease outbreaks [60, 61]. The low infection rates in people, although likely to be a gross underestimate [62], suggest low infection rates in mosquitoes, decreasing the chances of arboviral detection in them. It would also mean fewer opportunities for infected urban mosquitoes to feed on peridomestic AGMs (‘spillback’ infection from people to NHPs) and in the process provide a source of virus for a sylvatic cycle as has been similarly investigated in Brazil [63,64,65,66,67].
We also found no infected mosquitoes in the more rural areas and rainforest where AGMs are more frequent. This is consistent with our finding that AGMs from these areas were seronegative and indicates that there is no sylvatic cycle on the island. In Africa and Asia, only sylvatic Aedes spp. are thought to play a role in the maintenance of sylvatic cycles of DENV, CHIKV and ZIKV. The only mosquito species we found in non-urban areas that is known to carry these viruses in a sylvatic setting was Ae. aegypti (Additional file 4: Table S2). DENV was isolated from these mosquitoes captured in forest galleries of Senegal in 1999 [21]. The Ae. aegypti mosquitoes we found on St Kitts (257/443; 58%) showed no morphological differences to those captured from urban environments and most likely represent ‘rewilding’. The rewilding was most likely driven by opportunistic use of more varied non-urban oviposition sites found in the Caribbean such as rock holes, calabashes, tree holes, leaf axils, bamboo joints, papaya stumps, coconut shells, bromeliads, ground pools, coral rock holes, crab holes and conch shells [68, 69]. The presence of Ae. aegypti in diverse habitats on St. Kitts suggests alternate non-human vertebrate blood-meal sources are available to them.
Despite high host preference of Ae. aegypti for humans, this species of mosquito will bite other available mammals. In Grenada, 28% (9/32) of Ae. aegypti blood meals were from non-human mammals (mongooses, domestic dogs and cats) [70] and similarly, on Puerto Rico, up to 21% (42/199) of Ae. aegypti blood meals were from domestic dogs [71]. Additionally, Ae. aegypti will feed on both NHPs and people where they live in close proximity as is the case on St. Kitts. For example, on the tourist island of Koh Chang in Thailand sampling mosquitoes in urban, forested and periurban areas of the national park revealed that 70% (21/30) of Ae. aegypti had fed on both people and NHPs suggesting less selective feeding behaviour in these situations [72].
Our blood-meal analysis showed that only a small percentage (3%) of the blood-fed mosquitoes we trapped had evidence of having fed on humans. We confirmed that the generalist feeder Ae. taeniorhynchus (and probably Culex spp.) will feed on humans and, interestingly, in Puerto Rico this species has been proven to feed on NHPs [73]. Using the specific HMBS gene sequence we determined for AGMs (BankIt2363830 AGM_seq MT742560) we found no evidence that the mosquitoes we trapped had fed on AGMs. This might be because there is a greater choice of vertebrate hosts (birds, mammals, reptiles and amphibians) in the rainforest and hence no feeding preference for AGMs. It could also be because we trapped mosquitoes only at ground level while sylvatic mosquitoes, more active at night [21, 43], might be more prevalent in the forest canopy where AGMs sleep.
Conclusions
Overall, we believe we have sufficient evidence to discount the presence of sylvatic cycles of CHIKV, DENV and ZIKV on St. Kitts. Using criteria commonly used in similar studies [4], mainly seropositivity and infected mosquitoes, we found no evidence that AGMs were exposed to the viruses or that mosquitoes on the island were infected.
Availability of data and materials
The data supporting the conclusions of this article are included within the article and its additional files.
Abbreviations
- AGM:
-
African green monkey
- BSF:
-
Behavioral Science Foundation
- CARPHA:
-
Caribbean Public Health Agency
- CHIKV:
-
Chikungunya virus
- CNR:
-
French National Centre for Arboviruses
- DENV:
-
Dengue virus
- NHP:
-
Non-human primate
- PAHO:
-
Pan American Health Organisation
- PRNT:
-
Plaque reduction neutralisation test
- RUSVM:
-
Ross University School of Veterinary Medicine
- ZIKV:
-
Zika virus
References
Bhatt S, Gething P, Brady O, Messina J, Farlow A, Moyes C. The global distribution and burden of dengue. Nature. 2012;496:504–7.
Sam IC, Chua CL, Rovie-Ryan JJ, Fu JYL, Tong C, Sitam FT, et al. Chikungunya virus in macaques. Malaysia. Emerg Infect Dis. 2015;21:1683–5.
Vasilakis N, Weaver SC. Flavivirus transmission focusing on Zika. Curr Opin Virol. 2018;22:30–5.
Valentine MJ, Murdock CC, Kelly PJ. Sylvatic cycles of arboviruses in non-human primates. Parasit Vectors. 2019;12:463.
Vasilakis N, Cardosa J, Hanley KA, Holmes EC, Weaver SC. Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat Rev Microbiol. 2011;9:532–41.
Wang E, Ni H, Xu R, Barrett AD, Watowich SJ, Gubler DJ, et al. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol. 2000;74:3227–34.
Diallo M, Thonnon J, Traore-Lamizana M, Fontenille D. Vectors of chikungunya virus in Senegal: current data and transmission cycles. Am J Trop Med Hyg. 1999;60:281–6.
Hanley KA, Monath TP, Weaver SC, Rossi SL, Richman RL, Vasilakis N. Fever versus fever: the role of host and vector susceptibility and interspecific competition in shaping the current and future distributions of the sylvatic cycles of dengue virus and yellow fever virus. Infect Genet Evol. 2013;19:292–311.
Tsetsarkin KA, Chen R, Weaver SC. Interspecies transmission and chikungunya virus emergence. Curr Opin Virol. 2016;16:143–50.
Althouse BM, Vasilakis N, Sall AA, Diallo M, Weaver SC, Hanley KA. Potential for Zika virus to establish a sylvatic transmission cycle in the Americas. PLoS Negl Trop Dis. 2016;10:e0005055.
Althouse BM, Guerbois M, Cummings DAT, Diop OM, Faye O, Faye A, et al. Role of monkeys in the sylvatic cycle of chikungunya virus in Senegal. Nat Commun. 2018;9:1046.
Weaver SC, Vasilakis N. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol. 2013;2009:523–40.
Mavian C, Dulcey M, Munoz O, Salemi M, Vittor AY, Capua I. Islands as hotspots for emerging mosquito-borne viruses: a one-health perspective. Viruses. 2019;11:1–28.
Pan American Health Organisation: PLISA health information platform for the Americas. https://www.paho.org/hq/index.php?option=com_topics&view=rdmore&cid=5927&item=chikungunya&type=statistics&Itemid=40931&lang=en. Accessed 6 Aug 2019.
Pan American Health Organisation: PLISA health information platform for the Americas. https://www.paho.org/hq/index.php?option=com_content&view=article&id=12390:zika-cumulative-cases&Itemid=42090&lang=en. Accessed 6 Aug 2019.
Mohammed H, Hayden MH, Lee E, Santiago LM, Krecek RC, Revan F, et al. Dengue in the campus community of an overseas American university: a cross-sectional study. J Infect Dev Ctries. 2019;13:233–9.
Gay N, Rousset D, Huc P, Matheus S, Ledrans M, Rosine J, et al. Seroprevalence of Asian lineage chikungunya virus infection on Saint Martin Island, 7 months after the 2013 emergence. Am J Trop Med Hyg. 2016;94:393–6.
Simmons G, Brès V, Lu K, Liss NM, Brambilla DJ, Ryff KR, et al. High incidence of chikungunya virus and frequency of viremic blood donations during epidemic, Puerto Rico, USA, 2014. Emerg Infect Dis. 2016;22:1221–8.
Hills SL, Fischer M, Petersen LR. Epidemiology of Zika virus infection. J Infect Dis. 2017;216:S868–74.
Valentine MJ, Ciraola B, Jacobs GR, Arnot C, Patrick J, Murdock CC. Effects of seasonality and land use on the abundance and distribution of mosquitoes on St. Kitts, West Indies. bioRxiv. 2020;089037.
Diallo M, Ba Y, Sall AA, Diop OM, Ndione JA, Mondo M, et al. Amplification of the sylvatic cycle of dengue virus type 2, Senegal, 1999–2000: entomologic findings and epidemiologic considerations. Emerg Infect Dis. 2003;9:362–7.
Dick GWA, Kitchen SF, Haddow AJ. Zika virus. Trans R Soc Trop Med Hyg. 1952;46:509–20.
McCrae AWR, Henderson BW, Kirya BG, Sempala SD. Chikungunya virus in the Entebbe area of Uganda. Trans R Soc Trop Med Hyg. 1971;65:152–61.
Rudnick A. Dengue virus ecology in Malaysia. Inst Med Res Malays Bull. 1986;23(23):51–2.
Dore KM, Gallagher CA, Mill A. Estimation of green monkey (Chlorocebus sabaeus) population size on the West Indian island of St. Kitts using telemetry and home range analysis. In press. 2020
Hamilton CM, Katzer F, Beierschmitt A, Soto E, Innes EA, Kelly PJ. First report of Toxoplasma gondii seroprevalence in wild-caught Caribbean African green monkeys. Parasit Vectors. 2014;7:571.
Scallan EM, Sample SH, Beierschmitt AM, Palmour RM. Hematologic and biochemical RIs for an aged population of captive African green monkeys (Chlorocebus aethiops sabaeus). Vet Clin Pathol. 2017;46:430–5.
Cawthon Lang KA. Primate factsheets: vervet (Chlorocebus) taxonomy, morphology & ecology. In: Primate Info Net. Wisconsin National Primate Research Center. https://primate.wisc.edu/primate-info-net/pin-factsheets/pin-factsheet-vervet-monkey. Accessed 30 Aug 2020.
Aliota MT, Dudley DM, Newman CM, Mohr EL, Gellerup DD, Breitbach ME, et al. Heterologous protection against Asian Zika virus challenge in Rhesus macaques. PLoS Negl Trop Dis. 2016;10:e0005168.
Lindsey HS, Calisher CH, Mathews JH. Serum dilution neutralization test for California group virus identification and serology. J Clin Microbiol. 1976;4:503–10.
Moreira-Soto A, de Carneiro O, Fischer C, Feldmann M, Kümmerer BM, Silva NS, et al. Limited evidence for infection of urban and peri-urban nonhuman primates with Zika and chikungunya viruses in Brazil. mSphere. 2018;3:e00523.
Belkin JN, Heinemann SJ. Collection Records of the Project “Mosquitoes of Middle America”. 4. Leeward Islands: Anguilla (ANG), Antigua (ANT), Barbuda (BAB), Monserrat (MNT), Nevis (NVS), St. Kitts (KIT). Mosq Syst. 1976;8:123–62.
Burkett-Cadena ND. Mosquitoes of the Southeastern United States. 1st ed. Tuscaloosa: The Univesrity of Alabama Press; 2013.
Darsie RF, Ward RA. Identification and Geographical Distribution of the mosquitoes of North America, North of Mexico. 2nd ed. Gainesville: University Press of Florida; 2005.
Wei L, Kelly P, Zhang J, Yi Y, Wang C. Use of a universal hydroxymethylbilane synthase (HMBS)-based PCR as an endogenous internal control and to enable typing of mammalian DNAs. Appl Microbiol Biotechnol. 2014;98:5579–87.
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–41.
Mohammed H, Evanson J, Revan F, Lee E, Krecek RC, Smith J. A mosquito survey of the twin-island Caribbean Nation of Saint Kitts and Nevis, 2010. J Am Mosq Control Assoc. 2015;31:360–3.
Apandi Y, Nazni WA, Azleen ZAN, Vythilingam I, Noorazian MY, Azahari AH, et al. The first isolation of chikungunya virus from non-human primates in Malaysia. J Med Entomol. 2009;1:35–9.
Buechler CR, Bailey AL, Weiler AM, Barry GL, Jasinska AJ, Freimer NB, et al. Prevalence of Zika virus infection in wild African primates. mSphere. 2017;2:e00392.
Eastwood G, Sang RC, Guerbois M, Taracha ELN, Weaver SC. Enzootic circulation of chikungunya virus in East Africa: Serological evidence in non-human Kenyan primates. Am J Trop Med Hyg. 2017;97:1399–404.
Inoue S, Morita K, Matias RR, Tuplano JV, Resuello RRG, Candelario JR, et al. Distribution of three arbovirus antibodies among monkeys (Macaca fascicularis) in the Philippines. J Med Primatol. 2003;32:89–94.
Kading RC, Borland EM, Cranfield M, Powers AM. Prevalence of antibodies to alphaviruses and flaviviruses in free-ranging game animals and nonhuman primates in the Greater Congo basin. J Wildl Dis. 2013;49:587–99.
Diallo D, Sall AA, Buenemann M, Chen R, Faye O, Diagne CT, et al. Landscape ecology of sylvatic chikungunya virus and mosquito vectors in southeastern Senegal. PLoS Negl Trop Dis. 2012;6:e1649.
Diallo D, Chen R, Diagne CT, Ba Y, Dia I, Sall AA, et al. Bloodfeeding patterns of sylvatic arbovirus vectors in southeastern Senegal. Trans R Soc Trop Med Hyg. 2013;107:200–3.
Dohoo I, Martin W, Stryhn H. Sampling. In: Dohoo I, Martin W, Stryhn H, editors. Methods Epidemiol Res. Charlottetown: VER Publishing Inc.; 2012. p. 48–55.
Sergeant E. Epitools Epidemiological Calculators. AusVet. 2018. http://epitools.ausvet.com.au. Accessed 12 June 2019.
Hickey AC, Koster JA, Thalmann CM, Hardcastle K, Tio PH, Cardosa MJ, et al. Serotype-specific host responses in Rhesus macaques after primary dengue challenge. Am J Trop Med Hyg. 2013;89:1043–57.
McGee CE, Lewis MG, Claire MS, Wagner W, Lang J, Guy B, et al. Recombinant chimeric virus with wild-type dengue 4 virus premembrane and envelope and virulent yellow fever virus asibi backbone sequences is dramatically attenuated in nonhuman primates. J Infect Dis. 2008;197:693–7.
Meng W, Li L, Xiong W, Fan X, Deng H, Bett AJ, et al. Efficient generation of monoclonal antibodies from single rhesus macaque antibody secreting cells. MAbs. 2015;7:707–18.
Moi ML, Ami Y, Shirai K, Lim CK, Suzaki Y, Saito Y, et al. Formation of infectious dengue virus-antibody immune complex in vivo in marmosets (Callithrix jacchus) after passive transfer of anti-dengue virus monoclonal antibodies and infection with dengue virus. Am J Trop Med Hyg. 2015;92:370–6.
Beck C, Jimenez-Clavero MA, Leblond A, Durand B, Nowotny N, Leparc-Goffart I, et al. Flaviviruses in europe: complex circulation patterns and their consequences for the diagnosis and control of West Nile disease. Int J Environ Res Public Health. 2013;10:6049–83.
De Silva AM, Dittus WPJ, Amerasinghe PH, Amerasinghe FP. Serologic evidence for an epizootic dengue virus infecting toque macaques (Macaca sinica) at Polonnaruwa, Sri Lanka. Am J Trop Med Hyg. 1999;60:300–6.
Kato F, Ishida Y, Kawagishi T, Kobayashi T, Hishiki T, Miura T, et al. Natural infection of cynomolgus monkeys with dengue virus occurs in epidemic cycles in the Philippines. J Gen Virol. 2013;94:2202–7.
Vour’c HG, Halos L, Desvars A, Boué F, Pascal M, Lecollinet S, et al. Chikungunya antibodies detected in non-human primates and rats in three Indian Ocean islands after the 2006 ChikV outbreak. Vet Res. 2014;45:52.
Beck C, Lowenski S, Durand B. Improved reliability of serological tools for the diagnosis of West Nile fever in horses within Europe. PLoS Negl Trop Dis. 2017;11:e0005936.
Hunsperger EA, McElroy KL, Bessoff K, Colón C, Barrera R, Muñoz-Jordán JL. West Nile virus from blood donors, vertebrates, and mosquitoes, Puerto Rico, 2007. Emerg Infect Dis. 2009;15:1298–300.
Bolfa P, Jeon I, Loftis A, Leslie T, Marchi S, Sithole F, et al. Detection of West Nile virus and other common equine viruses in three locations from the Leeward Islands, West Indies. Acta Trop. 2017;174:24–8.
White S, Lednicky J, Okech BA, Morris JG Jr, Dunford JC. Spondweni virus in field-caught Culex quinquefasciatus mosquitoes, Haiti, 2016. Emerg Infect Dis. 2018;24:1765–7.
Ochieng C, Lutomiah J, Makio A, Koka H, Chepkorir E, Yalwala S, et al. Mosquito-borne arbovirus surveillance at selected sites in diverse ecological zones of Kenya; 2007–2012. Virol J. 2013;10:140.
Ramírez AL, Van Den Hurk AF, Meyer DB, Ritchie SA. Searching for the proverbial needle in a haystack: advances in mosquito-borne arbovirus surveillance. Parasit Vectors. 2018;11:320.
Gu W, Novak RJ. Short report: Detection probability of arbovirus infection in mosquito populations. Am J Trop Med Hyg. 2004;71:636–8.
ten Bosch QA, Clapham HE, Lambrechts L, Duong V, Buchy P, Althouse BM, et al. Contributions from the silent majority dominate dengue virus transmission. PLoS Pathog. 2018;14:e1006965.
De Abreu FVS, Ferreira-De-Brito A, De Souza Azevedo A, Linhares JHR, De Oliveira Santos V, Miranda EH, et al. Survey on non-human primates and mosquitoes does not provide evidences of spillover/spillback between the urban and sylvatic cycles of yellow fever and Zika viruses following severe outbreaks in southeast Brazil. Viruses. 2020;12:364.
Favoretto SR, Araujo DB, Duarte NFH, Oliveira DBL, da Crus NG, Mesquita F, et al. Zika virus in peridomestic neotropical primates. Northeast Brazil. Ecohealth. 2019;16:61–9.
Fernandes RS, Bersot MI, Castro MG, Telleria EL, Ferreira-de-Brito A, Raphael LM, et al. Low vector competence in sylvatic mosquitoes limits Zika virus to initiate an enzootic cycle in South America. Sci Rep. 2019;9:20151.
Karna AK, Azar SR, Plante JA, Yun R, Vasilakis N, Weaver SC, et al. Colonized Sabethes cyaneus, a sylvatic new world mosquito species, shows a low vector competence for Zika virus relative to Aedes aegypti. Viruses. 2018;10:434.
Terzian ACB, Zini N, Sacchetto L, Rocha RF, Parra MCP, Del Sarto JL, et al. Evidence of natural Zika virus infection in neotropical non-human primates in Brazil. Sci Rep. 2018;8:16034.
Chadee DD, Ward RA, Novak RJ. Natural habitats of Aedes aegypti in the Caribbean - a review. J Am Mosq Control Assoc. 1998;14:5–11.
Powell JR, Tabachnick WJ. History of domestication and spread of Aedes aegypti - a review. Mem Inst Oswaldo Cruz. 2013;108:11–7.
Fitzpatrick DM, Hattaway LM, Hsueh AN, Ramos-Niño ME, Cheetham SM. PCR-based bloodmeal analysis of Aedes aegypti and Culex quinquefasciatus (Diptera: Culicidae) in St. George Parish, Grenada. J Med Entomol. 2019;56:1170–5.
Barrera R, Bingham AM, Hassan HK, Amador M, Mackay AJ, Unnasch TR. Vertebrate hosts of Aedes aegypti and Aedes mediovittatus (Diptera : Culicidae) in Rural Puerto Rico. J Med Entomol. 2012;49:917–21.
Khaklang S, Kittayapong P. Species composition and blood meal analysis of mosquitoes collected from a tourist island, Koh Chang, Thailand. J Vector Ecol. 2014;39:448–52.
Hemme RR, Poole-Smith BK, Hunsperger EA, Felix GE, Horiuchi K, Biggerstaff BJ, et al. Non-human primate antibody response to mosquito salivary proteins: implications for dengue virus transmission in Puerto Rico. Acta Trop. 2016;164:369–74.
Acknowledgements
This project was only possible because of the assistance of Gilbert Gordon, Michelle Evans, Mike Newberry, David Pecor, Heather Hotchin, Anna Becker, Julie Graves, Rob Gilbert, Kathleen Gilbert, Moses Humphreys, Emily Dodd, Hannah Mrozinski, Alix Saveedra, Meredith Nugent, Kate Huff, Rudel Williams, Emily Cutolo, Katie Chan, Elisa Loke, Sarah Scott, Daniel Martin, Laura McPherson, Ligia Pentzkelemus, Holly Bates, Bundi Wilde, Eva Bhatt, Alexis Cataldo, Jake Byrne, Josh Patterson, Gretchen Yinger, Heidy Pardo, Renee Pageauvega, Calla Reilly, Alejandra Ramirez, Sofia Simone, Kayla Smith, Susan Naibkhyl, Jaydenice Gomeziriza, Naikesha Daniels, Lohit Busanelli, Katalina Cruz, Charlie Arnot, Scott Huang and Dana Vanlandingham.
Funding
The project was funded by National Institute of Allergy and Infectious Diseases grant number 1R21AI128407-01 and the Ross University School of Veterinary Medicine.
Author information
Authors and Affiliations
Contributions
MJV and BC collected, identified and tested the mosquitoes for arboviruses and analysed their blood meals. MJV, BC, CG and KD captured AGMs for phlebotomy. BT, SM, BC, MA and MJV tested AGM sera. XL and IG advised on serology. CW advised on blood-meal analysis. AB, CG and TC advised on AGM capture and supplied AGM sera. MV supervised and advised on all laboratory analyses. PK and CCM designed and supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study methods were approved by the Institutional Animal Care and Use Committees (IACUC) of Ross University School of Veterinary Medicine (RUSVM) (IACUC 10.17.47Kelly), the Behavioral Research Foundation (BSF), St. Kitts and St. Kitts Biomedical Research Foundation with Virscio, Inc., St. Kitts. BSF is accredited by the Canadian Association of Animal Care and St. Kitts Biomedical Research Foundation is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Both facilities operate in compliance with The Guide for the Care and Use of Laboratory Animals under National Institute of Health (NIH) Office of Laboratory Animal Welfare (OLAW) guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Text S1.
AGM capture and phlebotomy.
Additional file 2: Text S2.
Blood-meal analysis.
Additional file 3: Table S1.
Counts of mosquito species per month on St Kitts from November 2017 to March 2019 with the wet season highlighted in grey (May-November).
Additional file 4: Table S2.
Counts of mosquito species caught across the five different land covers from November 2017 to March 2019 on St Kitts.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Valentine, M.J., Ciraola, B., Aliota, M.T. et al. No evidence for sylvatic cycles of chikungunya, dengue and Zika viruses in African green monkeys (Chlorocebus aethiops sabaeus) on St. Kitts, West Indies. Parasites Vectors 13, 540 (2020). https://doi.org/10.1186/s13071-020-04419-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04419-1