Abstract
Background
Apicomplexan parasites of the genus Cryptosporidium infect a wide range of animal species as well as humans. Cryptosporidium spp. can cause life threatening diarrhea especially in young animals, children, immunocompromised patients and malnourished individuals. Asymptomatic cryptosporidial infections in animals can also occur, making these animals potential reservoirs of infection.
Methods
In the present study, a molecular survey of Cryptosporidium spp. in ruminants that were slaughtered for human consumption in Yazd Province, located in central Iran was conducted. Faeces were collected per-rectum from 484 animals including 192 cattle, 192 sheep and 100 goats. DNA was extracted from all samples and screened for Cryptosporidium by PCR amplification of the 18S rRNA gene. Positives were Sanger sequenced and further subtyped by sequence analysis of the 60 kDa glycoprotein (gp60) locus.
Results
In total, Cryptosporidium spp. were detected in 22 animals: C. andersoni and C. bovis in seven and two cattle faecal samples, respectively, C. ubiquitum in five sheep, and C. xiaoi in six sheep and two goat samples, respectively. To our knowledge, this study provides for the first time, molecular information concerning Cryptosporidium species infecting goats in Iran, and is also the first report of C. ubiquitum and C. xiaoi from ruminants in Iran.
Conclusion
The presence of potentially zoonotic species of Cryptosporidium in ruminants in this region may suggest that livestock could potentially contribute to human cryptosporidiosis, in particular among farmers and slaughterhouse workers, in the area. Further molecular studies on local human populations are required to more accurately understand the epidemiology and transmission dynamics of Cryptosporidium spp. in this region.
Similar content being viewed by others
Background
Parasites of the genus Cryptosporidium are ubiquitous zoonotic pathogens of humans and animals and are responsible for significant number of water-borne outbreaks worldwide [1, 2]. Cryptosporidium spp. infect a wide range of mammals and also birds, amphibians, fishes and reptiles [3]. Of the 39 valid species, over 20 Cryptosporidium species and genotypes have been identified in human patients causing asymptomatic or mild to severe gastrointestinal disease. Of these, C. parvum and C. hominis are by far the most common etiological agents responsible for cryptosporidiosis in humans worldwide [4,5,6]. It has been shown that several Cryptosporidium species are not host-specific and can infect a wide host range [7].
In Iran, various studies have reported Cryptosporidium in humans [8], cattle and calves [9], sheep and goats [10], water buffaloes [11], camels [12], dogs [13], cats [14], horses [15], birds [16, 17], rodents [18], vegetables [19], wastewater [20] and recreational water [21]. There are also reports of simultaneous detection of Cryptosporidium in livestock and people associated with them such as farmers, shepherds and slaughterhouse workers, suggesting zoonotic transmission of Cryptosporidium spp. from animals to humans [22,23,24,25]. However, most of the studies focused on humans and animals with diarrhea. Although livestock can play a major role as a source of human cryptosporidiosis, not all of the infected livestock show clinical signs such as diarrhea [26]. Moreover, cross-contamination of raw meat with animal excreta in the process of slaughtering is a risk factor for human cryptosporidiosis [27, 28]. To date, there is no information about infection of ruminants with Cryptosporidium in Yazd Province, Iran. Therefore, the aim of this study was to use molecular tools to characterize Cryptosporidium spp. in livestock (sheep, goats and cattle) at a local abattoir.
Methods
Study area
Yazd Province (32.1006°N, 54.4342°E) is located in the center of Iran, in the heart of the Dasht-e Kavir desert. Yearly rainfall of less than 100 mm and frequent summer temperatures of above 40 °C has made it one of the driest major regions in Iran. With about 152,000 cattle, 406,000 sheep, 406,000 goats and 16,000 camels, this province has one of the lowest population of livestock in the country [29]. Intensive farming is common although there are few grazing areas for small ruminants in this region. Two major sources of water for raising livestock in the region are underground aquifers and urban water supply network. Prior to the present study, no information was available concerning Cryptosporidium species in cattle, sheep and goats in this province.
Collection of samples
From June to November 2017, faecal samples were collected per-rectum from 484 slaughtered animals at the Moein Dam industrialized abattoir. The sample size for the present study (192 cattle, 192 sheep and 100 goats) was calculated using EpiTools [30], based on the most recent enumeration of livestock in the region and published articles on the prevalence of Cryptosporidium in neighboring areas. Metadata such as age, sex, health status of animals and faecal consistency for each sample was recorded systematically. Samples were collected into individual plastic containers, placed in polystyrene foam containers beside dry ice storage boxes and shipped to Shahid Sadoughi University of Medical Sciences and Health Services, Yazd, Iran within few hours.
DNA extraction
Faeces were thoroughly homogenized using disposable wooden applicators, three freeze/thaw cycles were preformed, and the homogenates were vortexed with sterile glass beads. Genomic DNA (gDNA) was extracted using a DNA Blood and Tissues® extraction kit (MBST, Tehran, Iran). The DNA yield was assessed using a Thermo Scientific™ NanoDrop™ spectrophotometer (Thermo Fisher Scientific, Massachusetts, USA).
PCR amplification
All samples were screened for the presence of Cryptosporidium spp. at the 18S rRNA locus using a nested-PCR producing a ~ 611-bp product [31], with modified PCR conditions as previously described [32]. All samples positive for Cryptosporidium spp. at the 18S locus were further subtyped at the 60 kDa glycoprotein (gp60) locus using a nested-PCR producing a ~ 948-bp secondary product, as previously described [33].
Sequencing and molecular analysis
The amplified DNA from secondary PCR products was separated by gel electrophoresis and sent for sequencing using an ABI 3730XL DNA Analyzer at the Bioneer Company (Daejeon, Republic of Korea). Sanger sequencing chromatogram files were imported into Geneious Pro 8.1.6 [34], edited, analyzed, and aligned with reference sequences from GenBank using Clustal W (http://www.clustalw.genome.jp).
Statistical analyses
The prevalence of Cryptosporidium in faecal samples collected from each host species was expressed as the percentage of samples positive at the 18S locus, with 95% confidence intervals calculated assuming a binomial distribution, using the software Quantitative Parasitology 3.0 [35]. Chi-square and Fisher’s exact tests were performed using SPSS 20 for Windows (SPSS Inc. Chicago, USA), to determine if there was a statistical difference in the prevalence of Cryptosporidium between livestock species, their age and sex. P-values < 0.05 were considered statistically significant.
Results
Health status of animals and faeces consistency
Faecal samples were classified as non-formed faeces (pasty/watery; diarrheic) or formed faeces (normal; non-diarrheic). All of the faecal samples collected were formed (96.6%; 95% CI: 94.9–98.2%) with the exception of 15 (five diarrheal, four semi-liquid and six pasty). Of 194 cattle samples examined, one had mastitis and one was pregnant. All the sheep and goats appeared to be healthy.
Prevalence of Cryptosporidium
Based on identification of Cryptosporidium at the 18S rRNA locus, the overall prevalence in the examined animals was 4.5% (22/484; 95% CI: 2.62–6.38%) comprising a prevalence of 4.7% (9/192; 95% CI: 1.71–7.69%) in cattle, 5.7% (11/192; 95% CI: 2.42–8.97%) in sheep and 2% (2/100; 95% CI: 0–4.74%) in goats.
Of the 22 faecal samples positive for Cryptosporidium, the consistency of all the samples were formed. Among 261 male animals, 15 were infected with Cryptosporidium (5.7%; 95% CI: 2.89–8.50%), and from 221 female animals, seven were positive for Cryptosporidium (3.1%; 95% CI: 0.82–5.38%) however, no significant correlation between cryptosporidiosis and sex of the animals was found. Cryptosporidiosis was more prevalent in younger animals (P = 0.019). For the prevalence of Cryptosporidium in faecal samples from livestock categorized by livestock species, sex and age groups, see Additional file 1: Table S1 and Additional file 2: Table S2.
Cryptosporidium species and subtypes
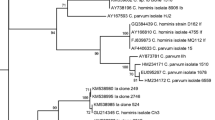
Sequence analysis at the 18S locus identified C. andersoni (n = 7) and C. bovis (n = 2) in cattle, C. ubiquitum (n = 5) and C. xiaoi (n = 6) in sheep and C. xiaoi (n = 2) in goats. Out of five samples positive for C. ubiquitum at the 18S locus, two were successfully subtyped at the gp60 locus (subtype family XIIa).
The sequences obtained from 22 livestock faeces in the present study were deposited in the GenBank database under the accession numbers MN153790–MN153794, MN394767–MN394783, MK797741 and MK801770 (Additional file 3: Table S3).
Discussion
In the present study, Cryptosporidium parasites were detected in the faeces of livestock that were slaughtered for human consumption in Yazd Province, central Iran, using molecular techniques. The prevalence was 4.5%, 5.7% and 2% in cattle, sheep and goats, respectively.
Since the first report of cryptosporidiosis in Iran in 1984 [36] numerous studies have reported different Cryptosporidium species in a wide range of mammalian hosts including C. parvum, C. muris, C. meleagridis, C. wrairi and C. hominis from humans [37,38,39,40], C. parvum from rats [41], C. baileyi from poultry and turkeys [9, 16], C. parvum from dogs [13], C. parvum, C. hominis, C. canis and Cryptosporidium pig genotype from recreational water [21, 42] and C. andersoni and C. xiaoi from livestock wastewater samples [20]. In Iran, it is estimated that 3.6% of children, 2.9% of healthy people, 1.3% of gastroenteritis patients and 4.5% of immunocompromised individuals are infected with Cryptosporidium species [8].
The present study provides the first insight into Cryptosporidium species infecting goats in Iran and is the first report of C. xiaoi in goats (prevalence of 2%, 95% CI: 0–4.74%). Worldwide, C. xiaoi and C. ubiquitum are responsible for over 90% of cryptosporidial infections in sheep and goats [7]. In Iran however, there is little information about caprine cryptosporidiosis, with all previous studies based on morphology alone, reporting prevalences ranging between 2.5–17.6% [43]. Studies in other countries have reported that 7.7% of healthy adult goats in Spain were infected with Cryptosporidium [44] and in France, a higher prevalence of Cryptosporidium (20%) was detected in goats aged 1–7 years in farms that had a previous history of diarrhoeal disease compared to farms that did not (6.6%) [45]. Cryptosporidium xiaoi has previously been reported in lambs, sheep, goats, yaks and kangaroos [46, 47] and in the present study, C. xiaoi was detected in adult goats aged 2 and 3 years-old, which is in contrast to previous reports of this species in goat kids [47, 48]. Further molecular-based studies are needed in Iran and other regions for better understanding of species infecting goats.
In sheep, C. xiaoi (n = 6) and C. ubiquitum (n = 5) were identified in the present study (11/192, 5.7%). These two species are responsible for over 90% of cryptosporidial infections in sheep and goats worldwide [7]. This is the first report of C. ubiquitum in Iran. Previous studies in sheep in Iran based on microscopy, have reported prevalences of 6.7–17.2% [49]. In the only molecular-based study, 22 (1.5%) of 1485 sheep faecal samples from Tehran city, were positive by microscopy and were typed as C. andersoni (n = 20) and C. parvum (n = 2) [50]. Cryptosporidium ubiquitum is a zoonotic parasite with a very wide host range [33, 51]. Infection of humans with C. ubiquitum has been reported primarily in industrialized nations and C. ubiquitum has also been detected in water sources, stormwaters and wastewaters in many geographical locations [33].
In the present study C. andersoni was detected in seven cattle faecal specimens (3.6% in 192; 95% CI: 0.98–6.23%) and C. bovis in two (1.04% in 192; 95% CI: 0.39–2.47%). In Iran, to date, C. parvum, C. bovis, C. andersoni, C. muris, C. wrairi, C. serpentis, and C. baileyi have been identified in calves and cattle using molecular tools [37, 39, 52,53,54]. Cryptosporidium parvum and C. bovis are the most commonly reported species infecting cattle worldwide, followed by C. ryanae and C. andersoni, with a few reports of C. occultus, C. ubiquitum and C. xiaoi [7]. However, previous studies on cattle in Iran, suggest that C. parvum and C. andersoni are the dominant species.
In this study cryptosporidiosis was more prevalent in younger animals. Age is often a significant variable for cryptosporidiosis status and etiology, and an age-related pattern in distribution of Cryptoposridium species has been observed in cattle, i.e. C. parvum is the predominant species in pre-weaned, monogastric calves up to 2 months of age, C. bovis and C. ryanae in older calves and young stock, whereas C. andersoni is mainly found in young stock and adult cattle [55]. In small ruminants however, species distribution differs between studies and between age groups within studies [55, 56]. Further studies in Iranian livestock of all ages including both symptomatic and asymptomatic individuals are required to better understand the associations between age, species/genotypes of Cryptosporidium and clinical outcome of cryptosporidiosis.
Faecal consistency of Cryptosporidium-infected adult ruminants in the present study was normal however, evidence supports an association between clinical disease and certain Cryptosporidium-species in livestock. In cattle, C. parvum often results in acute enteritis and pasty to watery diarrhea that in some cases lead to mortality from dehydration; C. andersoni is not associated with overt clinical signs but results in reduced milk production and weight gain; and infections with C. bovis and C. ryanae have not been associated with illness [56]. In small ruminants however, more research is needed to determine if species/genotype effects are significantly associated with production outcomes. Nevertheless, based on the current knowledge, C. parvum is more likely to be found in clinically ill lambs, whereas C. xiaoi and C. ubiquitum are more likely to be found in healthy lambs although the latter two species have been found in mild to severe diarrhoeal cases [55, 56].
In the present study C. ubiquitum, C. andersoni and C. bovis, all of which have zoonotic potential were found in adult ruminants. Studies suggest that the overall prevalence of Cryptosporidium declines with increasing age [57]. However, a study in England and Wales identified Cryptosporidium in 18.6% of adult cattle and 26.1% of adult sheep [58]. In Spain, Cryptosporidium prevalence in healthy adult cows was 8.4%, in sheep 5.3%, and in goats 7.7%, suggesting that asymptomatic adults can act as reservoirs for spreading oocysts in herds and pose a potential health risk for humans [44]. Although the prevalence of infection in the present study was low, the potential exists for contamination of water sources and for meat at slaughter and also abattoir wastewater, which is of public health concern.
Conclusions
To our knowledge, this study reports for the first time C. ubiquitum in Iran and C. xiaoi from ruminants in the country. The presence of zoonotic species C. ubiquitum, C. andersoni and C. bovis may suggest that livestock could contribute to human cryptosporidiosis, in particular among farmers and slaughterhouse workers, in the area. Further molecular studies in human populations is required to more accurately understand the epidemiology and transmission dynamics of Cryptosporidium spp. in this region.
Availability of data and materials
All data generated or analyzed during this study are included in this published article. The newly generated sequences were submitted to the GenBank database under the accession numbers MN153790-MN153794, MN394767-MN394783, MK797741 and MK801770.
References
Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks–an update 2004–2010. Water Res. 2011;45:6603–14.
Alsmark C, Nolskog P, Angervall AL, Toepfer M, Winiecka-Krusnell J, Bouwmeester J, et al. Two outbreaks of cryptosporidiosis associated with cattle spring pasture events. Vet Parasitol Reg Stud Rep. 2018;14:71–4.
Zahedi A, Paparini A, Jian F, Robertson I, Ryan U. Public health significance of zoonotic Cryptosporidium species in wildlife: critical insights into better drinking water management. Int J Parasitol Parasites Wildl. 2016;5:88–109.
Ryan U, Paparini A, Monis P, Hijjawi N. It’s official—Cryptosporidium is a gregarine: what are the implications for the water industry? Water Res. 2016;105:305–13.
Xiao L, Cama VA. Cryptosporidium and cryptosporidiosis. In: Ortega YR, Sterling CR, editors. Foodborne parasites. Cham: Springer; 2018. p. 73–117.
Ryan U, Zahedi A, Paparini A. Cryptosporidium in humans and animals—a one health approach to prophylaxis. Parasite Immunol. 2016;38:535–47.
Feng Y, Ryan UM, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34(11):997–1011.
Berahmat R, Spotin A, Ahmadpour E, Mahami-Oskouei M, Rezamand A, Aminisani N, et al. Human cryptosporidiosis in Iran: a systematic review and meta-analysis. Parasitol Res. 2017;116:1111–28.
Meamar AR, Guyot K, Certad G, Dei-Cas E, Mohraz M, Mohebali M, et al. Molecular characterization of Cryptosporidium isolates from humans and animals in Iran. Appl Environ Microbiol. 2007;73:1033–5.
Fasihi Harandi M, Fotouhi Ardakani R. Cryptosporidium infection on sheep and goats in Kerman: epidemiology and risk factor analysis. J Fac Vet Med Univ Tehran. 2008;63:47–51 (Article in Persian with English abstract).
Bahrami S, Alborzi AR, Molayan PH, Purbaram S, Mousavi B. prevalence of Cryptosporidium spp. infection and its association with diarrhea in buffalo calves in Khuzestan, a southwestern province of Iran. Buffalo Bull. 2014;33:393–9.
Sazmand A, Joachim A. Parasitic diseases of camels in Iran (1931–2017)—a literature review. Parasite. 2017;24:21.
Ranjbar R, Mirhendi H, Izadi M, Behrouz B, Mohammadi Manesh R. Molecular identification of Cryptosporidium spp. in Iranian dogs using seminested PCR: a first report. Vector Borne Zoonotic Dis. 2018;18:96–100.
Mirzaghavami M, Sadraei J, Forouzandeh M. Detection of Cryptosporidium spp. in free ranging animals of Tehran, Iran. J Parasit Dis. 2016;40:1528–31.
Ghadrdan AR, Hamidinejat H, Alizadehnia P. A Survey on frequency of equine cryptosporidiosis in Ahvaz. J Vet Clin Pathol. 2013;6:1723–7 (Article in Persian with English abstract).
Hamidinejat H, Jalali MHR, Jafari RA, Nourmohammadi K. Molecular determination and genotyping of Cryptosporidium spp. in fecal and respiratory samples of industrial poultry in Iran. Asian Pac J Trop Dis. 2014;7:517–20.
Radfar MH, Asl EN, Seghinsara HR, Dehaghi MM, Fathi S. Biodiversity and prevalence of parasites of domestic pigeons (Columba livia domestica) in a selected semiarid zone of South Khorasan, Iran. Trop Anim Health Prod. 2012;44:225–9.
Gholipoury M, Rezai HR, Namroodi S, Khazaeli FA. Zoonotic and non-zoonotic parasites of wild rodents in Turkman Sahra, northeastern Iran. Iran J Parasitol. 2016;11:350–7.
Fallah AA, Makhtumi Y, Pirali-Kheirabadi K. Seasonal study of parasitic contamination in fresh salad vegetables marketed in Shahrekord, Iran. Food Control. 2016;60:538–42.
Hatam-Nahavandi K, Mohebali M, Mahvi A-H, Keshavarz H, Najafian H-R, Mirjalali H, et al. Microscopic and molecular detection of Cryptosporidium andersoni and Cryptosporidium xiaoi in wastewater samples of Tehran Province, Iran. Iran J Parasitol. 2016;11:499–506.
Naeini KM, Asadi M, Chaleshtori MH. Detection and molecular characterization of Cryptosporidium species in recreational waters of Chaharmahal va Bakhtiyari Province of Iran using nested-PCR-RFLP. Iran J Parasitol. 2011;6:20–7.
Sazmand A, Rasooli A, Nouri M, Hamidinejat H, Hekmatimoghaddam S. Prevalence of Cryptosporidium spp. in camels and involved people in Yazd Province, Iran. Iran J Parasitol. 2012;7:80–4.
Nouri M, Toroghi R. Asymptomatic cryptosporidiosis in cattle and humans in Iran. Vet Rec. 1991;128:358–9.
Nouri M, Karami M. Asymptomatic cryptosporidiosis in nomadic shepherds and their sheep. J Infect. 1991;23:331–3.
Jafari R, Maghsood AH, Fallah M. Prevalence of Cryptosporidium infection among livestock and humans in contact with livestock in Hamadan district, Iran, 2012. J Res Health Sci. 2012;13:86–9.
Xiao L, Feng Y. Zoonotic cryptosporidiosis. FEMS Immunol Med Microbiol. 2008;52:309–23.
Budu-Amoako E, Greenwood SJ, Dixon BR, Barkema HW, McClure J. Foodborne illness associated with Cryptosporidium and Giardia from livestock. J Food Protect. 2011;74:1944–55.
Laberge I, Griffiths MW, Griffiths M. Prevalence, detection and control of Cryptosporidium parvum in food. Int J Food Microbiol. 1996;32:1–26.
Ministry of Agriculture-Jahad of Iran. Annual production report. Tehran; 2018 (in Persian).
Sergeant E. Epitools epidemiological calculators. 2018. http://epitools.ausvet.com.au/. Accessed 4 Sept 2019.
Silva SO, Richtzenhain LJ, Barros IN, Gomes AM, Silva AV, Kozerski ND, et al. A new set of primers directed to 18S rRNA gene for molecular identification of Cryptosporidium spp. and their performance in the detection and differentiation of oocysts shed by synanthropic rodents. Exp Parasitol. 2013;135:551–7.
Zahedi A, Phasey J, Boland T, Ryan U. First report of Cryptosporidium species in farmed and wild buffalo from the Northern Territory, Australia. Parasitol Res. 2016;115:1349–53.
Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, et al. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg Infect Dis. 2014;20:217.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9.
Rózsa L, Reiczigel J, Majoros G. Quantifying parasites in samples of hosts. J Parasitol. 2000;86:228–32.
Gharagozlou M. Report of one case of cryptosporidiosis in a calf and analysis of this disease. J Fac Vet Med Univ Tehran. 1984;40:81–9 (Article in Persian).
Azami M, Moghaddam DD, Salehi R, Salehi M. The identification of Cryptosporidium species in Isfahan, Iran by PCR-RFLP analysis of the 18S rRNA gene. Mol Biol. 2007;41:851–6.
Pirestani M, Sadraei J, Zavvar M, Vaeznia H. Molecular characterization of Cryptosporidium isolates from human and bovine using 18S rRNA gene in Shahriar county of Tehran, Iran. Parasitol Res. 2008;103:467–72.
Nazemalhosseini-Mojarad E, Haghighi A, Taghipour N, Keshavarz A, Mohebi SR, Zali MR, et al. Subtype analysis of Cryptosporidium parvum and Cryptosporidium hominis isolates from humans and cattle in Iran. Vet Parasitol. 2011;179:250–2.
Rafiei A, Rashno Z, Samarbafzadeh A, Khademvatan S. Molecular characterization of Cryptosporidium spp. isolated from immunocompromised patients and children. Jundishapur J Microbiol. 2014;7:e9183.
Bahrami F, Sadraei J, Frozandeh M. Molecular characterization of Cryptosporidium spp. in wild rats of Tehran, Iran using 18S rRNA gene and PCR-RFLP method. Jundishapur J Microbiol. 2012;2012:486–90.
Mohammadi Ghaleh Bin B, Fallah E, Asgharzadeh M, Kazemi A. Detection of Cryptosporidium species in water resources of Ardabil Province by PCR, RFLP. J Ardabil Univ Med Sci. 2007;7:177–83 (Article in Persian with English abstract).
Mohammadzadeh A. Study on the frequency of cryptosporidiosis in lambs of Ahwaz district of Khuzestan Province. DVM Thesis, Shahid Chamran University of Ahvaz; 1993 (in Persian).
Castro-Hermida JA, Almeida A, González-Warleta M, da Costa JMC, Rumbo-Lorenzo C, Mezo M. Occurrence of Cryptosporidium parvum and Giardia duodenalis in healthy adult domestic ruminants. Parasitol Res. 2007;101:1443–8.
Castro-Hermida J, Delafosse A, Pors I, Ares-Mazás E, Chartier C. Giardia duodenalis and Cryptosporidium parvum infections in adult goats and their implications for neonatal kids. Vet Rec. 2005;157:623–7.
Yang R, Fenwick S, Potter A, Ng J, Ryan U. Identification of novel Cryptosporidium genotypes in kangaroos from Western Australia. Vet Parasitol. 2011;179:22–7.
Fayer R, Santín M. Cryptosporidium xiaoi n. sp. (Apicomplexa: Cryptosporidiidae) in sheep (Ovis aries). Vet Parasitol. 2009;164:192–200.
Díaz P, Quílez J, Robinson G, Chalmers RM, Díez-Baños P, Morrondo P. Identification of Cryptosporidium xiaoi in diarrhoeic goat kids (Capra hircus) in Spain. Vet Parasitol. 2010;172:132–4.
Ahourai P, Ezzi A, Gholami M, Vandyoosefi J, Kargar R, Maalhagh N. Cryptosporidium spp. in new born lambs in Iran. Trop Anim Health Prod. 1985;17:6–8.
Dalimi A, Tahvildar F, Ghaffarifar F. Report of Cryptosporidium andersoni based on HSP70 gene amplification isolated from sheep in Tehran. J Vet Microbiol. 2017;13:19–27 (Article in Persian with English abstract).
Fayer R, Santín M, Macarisin D. Cryptosporidium ubiquitum n. sp. in animals and humans. Vet Parasitol. 2010;172:23–32.
Fotouhi Ardakani R, Fasihi Harandi M, Solayman Banai S, Kamyabi H, Atapour M, Sharifi I. Epidemiology of Cryptosporidium infection of cattle in Kerman/Iran and molecular genotyping of some isolates. J Kerman Univ Med Sci. 2008;15:313–20 (Article in Persian with English abstract).
Keshavarz A, Haghighi A, Athari A, Kazemi B, Abadi A, Mojarad EN. Prevalence and molecular characterization of bovine Cryptosporidium in Qazvin province, Iran. Vet Parasitol. 2009;160:316–8.
Mirzai Y, Yakhchali M, Mardani K. Cryptosporidium parvum and Cryptosporidium andersoni infection in naturally infected cattle of northwest Iran. Vet Res Forum. 2014;5:55–60.
Robertson LJ, Björkman C, Axén C, Fayer R. Cryptosporidiosis in farmed animals. In: Caccio SM, Giovanni W, editors. Cryptosporidium: parasite and disease. Vienna: Springer; 2014. p. 149–235.
Santín M. Clinical and subclinical infections with Cryptosporidium in animals. N Z Vet J. 2013;61:1–10.
Fayer R, Santín M, Trout JM. Prevalence of Cryptosporidium species and genotypes in mature dairy cattle on farms in eastern United States compared with younger cattle from the same locations. Vet Parasitol. 2007;145:260–6.
Smith R, Chalmers R, Mueller-Doblies D, Clifton-Hadley F, Elwin K, Watkins J, et al. Investigation of farms linked to human patients with cryptosporidiosis in England and Wales. Prev Vet Med. 2010;94:9–17.
Acknowledgements
The authors wish to thank Dr. Mehdi Komoripanah supervisor of Moein Dam Industrialized Abattoir.
Funding
This study was supported by Shahid Sadoughi University of Medical Sciences and Health Services, Yazd, Iran (under the framework of Mrs. Zohre Firoozi and Mrs. Narges Kiani M.Sc. theses), and Bu-Ali Sina University, Hamedan, Iran.
Author information
Authors and Affiliations
Contributions
Conceptualization: AS, FA, AZ and UR. Methodology: ZF, NK, AZ, AA, AF and BE. Statistical analyses: ZF and AD. Writing: original draft preparation: AS and ZF. Writing: review and editing: AZ and UR. Supervision: FA and UR. Project administration: FA. Funding acquisition: FA and AS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Faecal samples used for this study were collected from animals in Moein Dam slaughterhouse in accordance with the veterinary laws of I. R. Iran.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Prevalence of Cryptosporidium in livestock faecal samples by PCR at the 18S rRNA gene and in males and females.
Additional file 2: Table S2.
Prevalence of Cryptosporidium in livestock faecal samples by PCR at the 18S rRNA gene categorized by age.
Additional file 3: Table S3.
Cryptosporidium species in faeces of ruminants from Yazd, Iran.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Firoozi, Z., Sazmand, A., Zahedi, A. et al. Prevalence and genotyping identification of Cryptosporidium in adult ruminants in central Iran. Parasites Vectors 12, 510 (2019). https://doi.org/10.1186/s13071-019-3759-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3759-2