Abstract
Background
The important roles of microbial flora in tick biology and ecology have received much attention. Dermacentor marginatus and Dermacentor reticulatus are known vectors of various pathogens across Europe, including Slovakia. However, their bacterial microbiomes are poorly explored.
Methods
In this study, bacterial microbiomes of field-collected D. marginatus and D. reticulatus from Slovakia were characterized using 16S rRNA high-throughput sequencing.
Results
Different analyses demonstrated that the D. marginatus and D. reticulatus microbiomes differ in their diversity and taxonomic structures. Furthermore, species- and sex-specific bacteria were detected in the two species. A possible bacterial pathogen “Candidatus Rhabdochlamydia sp.” was detected from D. marginatus males. Among the observed bacteria, Rickettsia showed high abundance in the two species. Several maternally inherited bacteria such as Coxiella, Arsenophonus, Spiroplasma, Francisella and Rickettsiella, were abundant, and their relative abundance varied depending on tick species and sex, suggesting their biological roles in the two species.
Conclusions
The bacterial microbiomes of field-collected D. marginatus and D. reticulatus were shaped by tick phylogeny and sex. Maternally inherited bacteria were abundant in the two species. These findings are valuable for understanding tick-bacteria interactions, biology and vector competence of ticks.
Similar content being viewed by others
Background
Ticks are obligate blood-sucking parasitic arthropods, feeding on mammals, reptiles, birds and amphibians. More than 900 tick species have been identified worldwide, and many species are of great economic and epidemiological importance [1, 2]. Ticks carry and transmit various pathogens, including bacteria, viruses, protozoans and helminths [3]. Tick-borne diseases (TBDs) caused by these pathogens, such as human granulocytic anaplasmosis (HGA), Lyme disease, tick-borne encephalitis (TBE), babesiosis, etc., are distributed worldwide and resulting in serious harms [4]. Globally, tick-borne pathogens cause over 100,000 cases of human diseases yearly [1]. Every year about 65,000 people are infected in EU countries (https://ecdc.europa.eu/en/tick-borne-diseases). Habitat changes, climate changes, human activities and globalization are responsible for the emergence, spreading and migration of hosts, vectors, parasites and pathogens as well as for the rising incidence and diversity of vector-borne infections [5,6,7]. To date, at least 33 new tick-borne pathogens (TBPs) have been found in China [8]. Similarly, in Europe, climate change most predominantly affects seasonal range expansions and contractions of vector-borne diseases even in small areas [9]. For example, TBE cases moved from lowlands to sub mountainous areas in Slovakia since 1980, most likely because of rising temperature [10].
Given the importance of ticks as vectors of pathogens, aspects of tick biology and ecology have received much attention [2, 11, 12]. The tick microbiome comprises of communities of TBPs, viruses, bacteria and eukaryotes [13]. The rapid development of DNA and RNA sequencing platforms, especially high-throughput next-generation sequencing (NGS) technologies, have served as key drivers in our ability to realize the complexity of the tick microbiome in great detail [13, 14]. A series of studies have suggested that these non-pathogenic microorganisms are also abundant in ticks and have important roles in affecting tick biology and pathogen transmission [4, 13, 15,16,17,18,19]. A typical example is Coxiella-like endosymbiont, which has been reported as essential for tick survival and reproduction in Amblyomma americanum [20], Haemaphysalis longicornis [21] and Rhipicephalus microplus [22]. Recently, empirical evidence of an obligate B vitamin provisioning symbiont in ticks was found [23]. Non-pathogenic microorganisms also influence pathogens in different ways. For example, Ixodes scapularis fed on antibiotic-treated mice exhibited a modified gut microbiome, resulting in increased feeding and low Borrelia burgdorferi colonization rates [24]. Similarly, Gall et al. [15] found that a disrupted microbiome of Dermacentor andersoni is correlated with Anaplasma marginale and Francisella novicida susceptibility. These findings are paramount to fully exploiting the microbiome in order to control ticks and TBDs.
Dermacentor marginatus and Dermacentor reticulatus are two key tick vectors of various pathogens [1, 25, 26]. They are widespread in Europe, ranging from Portugal to Ukraine (and continue to the east of Kazakhstan) [27]. They are also distributed in China [25] and Russia [28]. Slovakia is located in central Europe; its climate lies between the temperate and continental climate zones with relatively warm summers and cold, cloudy and humid winters. The distributions and vector competences of D. marginatus and D. reticulatus have been fully investigated in Slovakia [29,30,31,32,33,34,35,36,37]. A survey found that D. reticulatus has extended its range in the surroundings of its former habitats [31]. In addition, the influences of global climate changes on the structures and dynamics of TBDs in mountain areas were assessed under a research project supported by governments of China and Slovakia.
It is evident that D. marginatus and D. reticulatus have great importance in medical and animal husbandry in Slovakia. Investigation of their microbiomes will aid in the control of ticks and TBDs. In this study, bacterial microbiomes of field-collected D. marginatus and D. reticulatus from Slovakia were characterized using 16S rRNA high-throughput sequencing.
Methods
Tick collection and sample preparation
Dermacentor marginatus and D. reticulatus were collected in the area of Slovak Karst, which is one of the mountain ranges of the Slovenské Rudohorie Mountains in the Carpathians in southern Slovakia. It consists of a complex of huge karst plains and plateaus. The area has been a protected landscape area since 1973, and in 2002, the Slovak Karst National Park was established. The park is also a UNESCO Biosphere Reserve and forms a UNESCO World Heritage site. Several endemic species of animals and plants live in this region, which has warm and moderately humid climate [38]. Tick collection sites were established in a small area situated on the northern grassy slope covered with scattered islands of xerophilous shrubs (212 meters above sea level, 48° 34′53.88″ N, 20° 46′ 44.43″ E), near the Hrhov village in eastern Slovakia. A small river and lake to the north and an old oak and hornbeam forest to the south surround the sampling site. These areas are usually used as the pastures for livestock grazing. Ticks were collected by the standard flagging method in the early spring of 2017 and were identified into developmental stages, species and sex using the taxonomic key [39]. Before study, ticks were stored at − 80 °C. A total of 48 adult ticks were used for this study (D. marginatus, n = 24; D. reticulatus, n = 24). For each species, according to tick sex, samples were grouped into three pools of 4 individuals each, and sample names are shown in Table 1.
DNA extraction
Prior to DNA extraction, ticks were surface-sterilized in three washes of 70% ethanol followed by one wash of sterile, nuclease-free, deionized water to avoid contamination from the environment. DNA extraction was performed using a QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany). The concentration and quality of DNA was measured using a Nanodrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA) and 1% gel electrophoresis detection, respectively.
16S rRNA PCR amplification and sequencing
The V3-V4 region of the bacterial 16S ribosomal RNA (rRNA) gene was amplified by PCR with barcode-indexed primers (338F: 5′-ACT CCT ACG GGA GGC AGC AG-3′ and 806R: 5′-GGA CTA CHV GGG TWT CTA AT-3′), using TransStart Fastpfu DNA Polymerase (TransGen, Beijing, China). PCRs were performed on GeneAmp® 9700 PCR instrument (Applied Biosystems, Foster City, CA, USA). This primer set, resulted in 420- to 460-bp PCR products. Amplicons were then purified by gel extraction (AxyPrep DNA GelExtraction Kit; Axygen Biosciences, Union City, CA, USA) and were quantified using QuantiFluor-ST (Promega, Madison, WI, USA). The purified amplicons were pooled in equimolar concentrations, and paired-end sequencing was performed on an Illumina MiSeq PE300 platform (Shanghai Majorbio Bio-pharm Technology Co., Ltd, Shanghai, China) using standard protocols.
Data analysis
The data were analyzed on the free online platform of Majorbio I-Sanger Cloud Platform (http://www.i-sanger.com). MiSeq sequence data were merged and filtered using the Trimmomatic software as previously described [40]. Quality-filtered merged reads were aligned to the Silva database [41] and screened for chimeras using Uchime algorithm [42]. Sequences with 97% similarity were then grouped into operational taxonomic units (OTUs) using OptiClust clustering algorithm [43]. The OTU table was processed in Qiime (MacQIIME v.1.9.0) [44]. OTUs were taxonomically assigned using the RDP Classifier v.2.2 [45] against the Greengenes 16S rRNA database v.13.5 with 70% confidence [46], and relative OTU abundances were summarized across taxonomic levels from domain to species.
Sufficient sequencing depth was determined based on rarefaction curves for observed number of OTUs from all samples. The bacterial composition of each sample was visualized as a bar figure. Sobs’ index and Shannon’s diversity index were calculated to measure bacterial community richness and diversity between groups, and Student’s t-test was used to test whether the two indices are significantly different. Analysis of similarities (ANOSIM) was used, with 999 permutations based on the Bray-Curtis index, to determine the percent variation of bacterial composition explained by tick species and sex. Beta diversity was examined using weighted and unweighted UniFrac analysis to compare the different groups and plotted in a principal coordinate analysis (PCoA). Wilcoxon rank-sum test was used to test for differences of bacterial composition between tick species and between males and females.
Results
MiSeq sequencing data
A total of 12 pooled samples were sequenced (Table 1), resulting in 1,045,584 raw reads. After filtration, 522,792 reads were generated and taxonomically assigned. The number of reads per sample was 30,632 to 65,290 (Additional file 1: Table S1). Rarefaction curves of the Shannonʼs index at OTU level indicated sufficient sequencing coverage, as demonstrated by observed Shannonʼs index accumulation curves reaching a plateau (Additional file 2: Figure S1).
Bacterial microbiome composition
In total, 550 OTUs were detected in 12 samples (Additional file 3: Table S2). The richness of the bacterial microbiome in D. reticulatus was higher than in D. marginatus, but the difference was not significant (t(10) = 1.1913, P = 0.084) (Fig. 1a). The diversity of the bacterial microbiome in D. reticulatus was significantly higher than in D. marginatus (t(10) = 3.757, P = 0.0037) (Fig. 1b). In D. marginatus, the bacterial microbiomes of females and males exhibited similar richness (t(4) = 0.2622, P = 0.81, Fig. 1a) and diversity (t(4) = 0.2527, P = 0.81, Fig. 1b) levels. However, the bacterial microbiome in male D. reticulatus had relatively higher richness (t(4) = 2.791, P = 0.049, Fig. 1a) and diversity (t(4) = 2.954, P = 0.042, Fig. 1b) compared with females.
Alpha diversity of bacterial microbiomes in Dermacentor marginatus and Dermacentor reticulatus. a Sobs index for each sample group. b Shannonʼs index for each sample group. *P < 0.05, **P < 0.01; ns indicates that differences between sample groups are not significant. Abbreviations: DmarF, female D. marginatus; DmarM, male D. marginatus; DretF, female D. reticulatus; DretM, male D. reticulatus
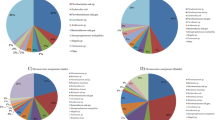
Bacterial microbiome was further assigned to 22 phyla, 40 classes, 89 orders, 161 families, 290 genera and 396 species. At the phylum level, Proteobacteria were the most dominant (60.4%), followed by Actinobacteria (25.76%), Chlamydiae (5.69%), Tenericutes (3.37%), Firmicutes (2.92%), Bacteroidetes (1.25%) and other phyla (0.63%). Most of bacterial phyla (17 of 22) were shared by D. marginatus and D. reticulatus, and by males and females within the same species. Chlamydiae were only found in male D. marginatus. Chlorobi and Parcubacteria were only detected in D. marginatus, while Ignavibacteriae, Armatimonadetes and an unclassified phylum were specific phyla in D. reticulatus (Fig. 2a, Additional file 3: Table S2). At the genus level, 218 bacterial genera were shared by D. marginatus and D. reticulatus. Among them, Rickettsia had the highest relative abundance (13.67%), followed by Brevibacterium (11.93%), “Candidatus Rhabdochlamydia” (9.3%), Pseudomonas (5.83%), Sphingomonas (5.36%), Methylobacterium (4.8%), Rhodococcus (3.83%) and Williamsia (3.68%). Of the 23 genera only detected in D. marginatus, Coxiella, Arsenophonus and Spiroplasma exhibited higher relative abundance (Fig. 2b). Two out of the 49 specific genera in D. reticulatus, Francisella and Rickettsiella had relatively high abundance (Fig. 2b).
Bacterial microbiome compositions of D. marginatus and D. reticulatus were significantly different according to ANOSIM (pseudo-R = 0.652, P = 0.003). Furthermore, PCoA analyses suggested that bacterial microbiome compositions were similar within the same tick species and the same tick sex (Fig. 3a, 3b).
Bacterial relative abundance differences
The relative abundance of the 15 top bacterial genera was compared using the Wilcoxon rank-sum test to detect possible differences. Five genera, i.e. Coxiella, Arsenophonus, Spiroplasma, Francisella and Rickettsiella, were detected at higher relative abundance (ranging between 6.7–26.2%). Except for “Candidatus Rhabdochlamydia” and Stenotrophomonas, most of the remaining genera had higher abundance in D. marginatus than that in D. reticulatus, and significant differences were found in the relative abundance of Williamsia and Staphylococcus (P < 0.05, Fig. 4a). In D. marginatus, females harbored more Coxiella, Spiroplasma and Stenotrophomonas. However, the relative abundance of Arsenophonus, Rickettsia and “Candidatus Rhabdochlamydia” were relatively high in males (Fig. 4b). In D. reticulatus, bacterial relative abundance differences between females and males were also observed, although the differences were not significant (Fig. 4c).
Discussion
In recent years, studies of the tick microbiome have been increasing, especially with the development and application of NGS technologies [14]. These studies have investigated the bacterial communities in different ticks [16, 47,48,49,50,51,52], and explored the influence of tick microbiomes on pathogen transmission and susceptibility [15, 16]; their findings strongly suggest that the bacterial microbiome has important roles in tick biology and ecology, and has a potential application in tick control.
To our knowledge, this study is the first to investigate the bacterial microbiomes of field-collected D. marginatus and D. reticulatus from Slovakia. The examined ticks were collected from the Slovak Karst region, of which the chosen study area (Hrhov) in particluar represents a significant biodiversity hotspot, not only in Slovakia but in the whole of central Europe. It is characterized by the presence of several endemic plant and animal species, and also by the co-occurrence of several tick species. In this area, in addition to the widely distributed Ixodes ricinus, tick species which are typical for the forest-steppe zones (D. marginatus and Haemaphysalis inermis) and the alluvial forests and wet meadows (D. reticulatus and Haemaphysalis concina) are also present. Moreover, the occurrence of Ixodes frontalis has been reported in this area [53]. Previous studies have found several pathogens in D. marginatus and D. reticulatus collected from Slovakia [34, 35, 37, 54]. In comparison, the bacterial microbiome of the two species is less known, and there are only two studies (on D. reticulatus in Russia [47] and D. marginatus in Turkey [48]).
MiSeq sequencing data generated from 12 pooled samples showed high quality and can be used for further analyses. The V3–V4 hypervariable regions of the 16S rRNA were amplified in this study, which is also used for microbiome surveys in ticks [16, 47] and in spider mites [55]. An earlier study by Sperling et al. [56] found that V4 amplicons can identify more bacteria in tick microbiome surveys.
Different analyses demonstrated that the D. marginatus and D. reticulatus microbiomes differ in their diversity and taxonomic structure. Furthermore, species- and sex-specific bacteria were detected from D. marginatus and D. reticulatus. In detail, D. reticulatus harbored more bacteria than D. marginatus, and the bacterial diversity in tick males seemed higher. The PcoA results suggested that the same species or sex have similar microbiome compositions. In addition, bacterial relative abundance differed between species and sexes, and specific bacteria were generally prevalent in their tick hosts. This study provides further evidence that host-related factors affect tick microbiome diversity and composition. Previous studies have revealed that the tick microbiome could vary depending on other factors, such as the season during which ticks were collected [57], geographical region [51, 58], tick developmental stages and tissues [16, 50, 58, 59], tick feeding status [60, 61] and presence of pathogens [17, 50].
Proteobacteria were the most abundant phylum in the two species and the phyla Actinobacteria, Bacteroidetes, Firmicutes and Tenericutes had high relative abundance; these findings are consistent with the findings in other tick species [60, 62, 63]. A special case was found in D. marginatus males, which had high relative abundance of Chlamydiae. These bacteria were further assigned to the order Chlamydiales, family Rhabdochlamydiaceae and “Candidatus Rhabdochlamydia”. Their 16S rRNA gene sequences were similar to “Candidatus Rhabdochlamydia porcellionis”, a known intracellular pathogen from the hepatopancreas of the terrestrial isopod Porcellio scaber [64]. Rhabdochlamydiaceae was also present in other arthropods, such as cockroaches [65] and dwarf spiders [66]. In ticks, Rhabdochlamydiaceae was identified in I. ricinus [67, 68] and Hyalomma dromedarii [67]. These observations suggest that arthropods can be reservoirs and vectors of the Rhabdochlamydiaceae. The pathogenic roles of Rhabdochlamydiaceae are not clear, mainly due to the almost complete absence of diagnostic tools and the difficulties encountered in attempts to cultivate Rhabdochlamydiaceae. Considering the fact that ticks can transmit some bacteria of Chlamydiales to humans and animals [69], investigating the prevalence of Chlamydiales within wild and farm animals, as well as the prevalence in humans with and without a history of tick bites, is necessary in the future.
As an obligate intracellular bacteria associated with ticks, Rickettsia can be divided into pathogens and non-pathogenic symbionts [70]. In the present study, Rickettsia has been shown to be prevalent in both D. marginatus and D. reticulatus. The 16S rRNA gene fragments used for amplification are highly conserved within Rickettsia, which hinders their species-level identification. Similar patterns of Rickettsia infection were found in D. marginatus studied in Turkey [48] and D. reticulatus studied in Russia [47]. Additionally, Duron et al. [19] found that Rickettsia-like endosymbionts are common in various tick species, including D. marginatus. The effects of non-pathogenic Rickettsia spp. on tick biology are poorly understood. D. marginatus and D. reticulatus are widely distributed across Europe and known as vectors of two pathogenic Rickettsia (R. slovaca and R. raoultii) [34, 71, 72]. Therefore, further efforts are needed to distinguish if Rickettsia are pathogenic or non-pathogenic endosymbionts and to explore their biological effects.
Besides the high prevalence of Rickettsia, some soil or environmental bacterial genera such as Brevibacterium, Pseudomonas, Sphingomonas and Rhodococcus were abundant in the two species of tick. These bacteria were also detected in many other tick species, although sterilization has been performed prior to DNA isolation [16, 24, 59, 62, 63]. This may be due to inadequate sterilization, or that these bacteria may have been ingested by ticks during feeding and therefore present in the tick midgut [14, 16]. Studies in nymphal and adult I. scapularis provided supportive evidence, as both dissected gut tissues and whole ticks showed many common genera such as Stenotrophomonas, Sphingobacterium, Pseudomonas and Acinetobacter, suggesting that these bacteria are likely bona fide tick gut residents [24, 59].
At least ten maternally inherited bacteria have been found in ticks [19]. Among them, five of six observed bacteria showed a specific association to tick species in this study. An earlier study by Duron et al. [19] revealed the presence of Coxiella, Rickettsia and Spiroplasma in D. marginatus, and the presence of Francisella in D. reticulatus. NGS analysis also found that Russian D. reticulatus harboured Francisella [47] whereas in another study of the bacterial infections of D. reticulatus in Slovakia, R. raoultii, R. slovaca, Coxiella burnetii, Coxiella-like and Francisella-like endosymbionts were detected [37]. NGS analysis of D. marginatus in Turkey only found Rickettsia [48]. These findings further suggest that the bacterial compositions in the two species are influenced by their geographical distribution. Tick sex is another factor influencing bacterial infections, as females and males had different bacterial abundance [47, 48]. The roles of most bacteria have yet to be clearly elucidated [18]. However, the essential roles of Coxiella-like and Francisella-like endosymbionts have been reported in several tick species, in which these bacteria may provide essential nutrients for the ticks [20,21,22,23]. Given the high prevalence of Coxiella-like and Francisella-like endosymbionts in D. marginatus and D. reticulatus, further studies examining the mutualistic relationships between these endosymbionts and their tick hosts are warranted. In addition, Spiroplasma and Arsenophonus were abundant in D. marginatus. Their presence in different tick species have also been summarized [73]. Spiroplasma and Arsenophonus act as male-killers in some other arthropod species [74, 75]. However, no male-killing effect was observed in D. marginatus, even though they were detected in males.
Conclusions
The bacterial microbiomes of field-collected D. marginatus and D. reticulatus from Slovakia differed in their diversity and taxonomic structure. Tick phylogeny and sex were two factors influencing the bacterial microbiome. In detail, D. reticulatus harbored more bacteria than D. marginatus, and the bacterial diversity in tick males seemed higher. A possible bacterial pathogen “Candidatus Rhabdochlamydia sp.” was detected from D. marginatus males. Rickettsia was the most abundant and other maternally inherited bacteria also had high relative abundance, although their biological roles are unclear. The occurrence of soil or environmental bacterial genera indicated that they may have been ingested by ticks during feeding. These findings will aid in the control of ticks and TBDs.
Availability of data and materials
All MiSeq sequencing data are available on the NCBI Sequence Read Archive (SRA) under the accession number PRJNA548395.
Abbreviations
- TBDs:
-
tick-borne diseases
- HGA:
-
human granulocytic anaplasmosis
- TBPs:
-
tick-borne pathogens
- NGS:
-
high-throughput next-generation sequencing
- 16S rRNA:
-
16S ribosomal RNA
- OUT:
-
operational taxonomic unit
References
de la Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci. 2008;13:6938–46.
Barker SC, Murrell A. Systematics and evolution of ticks with a list of valid genus and species names. In: Sonenshine DE, Roe RM, editors. Biology of ticks, vol. 2. New York: Oxford University Press; 2013. p. 1–39.
Jongejan F, Uilenber G. The global importance of ticks. Parasitology. 2004;129:S3–14.
de la Fuente J, Antunes S, Bonnet S, Cabezas-Cruz A, Domingos AG, Estrada-Peña A, et al. Tick-pathogen interactions and vector competence: identification of molecular drivers for tick-borne diseases. Front Cell Infect Microbiol. 2017;7:114.
Semenza JC, Suk JE. Vector-borne diseases and climate change: a European perspective. FEMS Microbiol Lett. 2017;365:fnx244.
Harrus S, Baneth G. Drivers for the emergence and re-emergence of vector-borne protozoal and bacterial diseases. Int J Parasitol. 2005;35:1309–18.
Gray JS, Dautel H, Estrada-Peña A, Kahl O, Lindgren E. Effects of climate change on ticks and tick-borne diseases in Europe. Interdiscip Perspect Infect Dis. 2009;2009:593232.
Fang LQ, Liu K, Li XL, Liang S, Yang Y, Yao HW, et al. Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect Dis. 2015;15:1467–79.
Semenza JC, Menne B. Climate change and infectious diseases in Europe. Lancet Infect Dis. 2009;9:365–75.
Lukan M, Bullova E, Petko B. Climate warming and tick-borne encephalitis, Slovakia. Emerg Infect Dis. 2010;16:524.
Randolph SE. Tick ecology: processes and patterns behind the epidemiological risk posed by ixodid ticks as vectors. Parasitology. 2004;129(Suppl. 1):S37–65.
Chmelař J, Kotál J, Karim S, Kopacek P, Francischetti IM, Pedra JH, et al. Sialomes and mialomes: a systems-biology view of tick tissues and tick-host interactions. Trends Parasitol. 2016;32:242–54.
Narasimhan S, Fikrig E. Tick microbiome: the force within. Trends Parasitol. 2015;31:315–23.
Greay TL, Gofton AW, Paparini A, Ryan UM, Oskam CL, Irwin PJ. Recent insights into the tick microbiome gained through next-generation sequencing. Parasit Vectors. 2018;11:12.
Gall CA, Reif KE, Scoles GA, Mason KL, Mousel M, Noh SM, et al. The bacterial microbiome of Dermacentor andersoni ticks influences pathogen susceptibility. ISME J. 2016;10:1846.
Zolnik CP, Prill RJ, Falco RC, Daniels TJ, Kolokotronis SO. Microbiome changes through ontogeny of a tick pathogen vector. Mol Ecol. 2016;25:4963–77.
Abraham NM, Liu L, Jutras BL, Yadav AK, Narasimhan S, Gopalakrishnan V, et al. Pathogen-mediated manipulation of arthropod microbiota to promote infection. Proc Natl Acad Sci USA. 2017;114:E781–90.
Bonnet SI, Binetruy F, Hernández-Jarguín AM, Duron O. The tick microbiome: why non-pathogenic microorganisms matter in tick biology and pathogen transmission. Front Cell Infect Microbiol. 2017;7:236.
Duron O, Binetruy F, Noël V, Cremaschi J, McCoy KD, Arnathau C, et al. Evolutionary changes in symbiont community structure in ticks. Mol Ecol. 2017;26:2905–21.
Zhong J, Jasinskas A, Barbour AG. Antibiotic treatment of the tick vector Amblyomma americanum reduced reproductive fitness. PLoS One. 2007;2:e405.
Zhang CM, Li NX, Zhang TT, Qiu ZX, Li Y, Li LW, et al. Endosymbiont CLS-Hl plays a role in reproduction and development of Haemaphysalis longicornis. Exp Appl Acarol. 2017;73:429–38.
Guizzo MG, Parizi LF, Nunes RD, Schama R, Albano RM, Tirloni L, et al. A Coxiella mutualist symbiont is essential to the development of Rhipicephalus microplus. Sci Rep. 2017;7:17554.
Duron O, Morel O, Noël V, Buysse M, Binetruy F, Lancelot R, et al. Tick-bacteria mutualism depends on B vitamin synthesis pathways. Curr Biol. 2018;28:1896–902.
Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, Pan J, et al. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe. 2014;15:58–71.
Yu Z, Wang H, Wang T, Sun W, Yang X, Liu J. Tick-borne pathogens and the vector potential of ticks in China. Parasit Vectors. 2015;8:24.
Földvári G, Široký P, Szekeres S, Majoros G, Sprong H. Dermacentor reticulatus: a vector on the rise. Parasit Vectors. 2016;9:314.
Rubel F, Brugger K, Pfeffer M, Chitimia-Dobler L, Didyk YM, Leverenz S, et al. Geographical distribution of Dermacentor marginatus and Dermacentor reticulatus in Europe. Ticks Tick-borne Dis. 2016;7:224–33.
Mediannikov O, Matsumoto K, Samoylenko I, Drancourt M, Roux V, Rydkina E, et al. Rickettsia raoultii sp. nov., a spotted fever group rickettsia associated with Dermacentor ticks in Europe and Russia. Int J Sys Evol Microbiol. 2008;58:1635–9.
Guryčová D, Kocianova E, Výrosteková V, Řeháček J. Prevalence of ticks infected with Francisella tularensis in natural foci of tularemia in western Slovakia. Euro J Epidemiol. 1995;11:469–74.
Špitalská E, Kocianova E. Detection of Coxiella burnetii in ticks collected in Slovakia and Hungary. Euro J Epidemiol. 2003;18:263–6.
Bullová E, Lukáň M, Stanko M, Peťko B. Spatial distribution of Dermacentor reticulatus tick in Slovakia in the beginning of the 21st century. Vet Parasitol. 2009;165:357–60.
Kubelova M, Tkadlec E, Bednář M, Roubalova E, Široký P. West-to-east differences of Babesia canis canis prevalence in Dermacentor reticulatus ticks in Slovakia. Vet Parasitol. 2011;180:191–6.
Majlathova V, Majlath I, Vichova B, Gul’ová I, Derdakova M, Sesztakova E, et al. Polymerase chain reaction confirmation of Babesia canis canis and Anaplasma phagocytophilum in dogs suspected of babesiosis in Slovakia. Vector Borne Zoonotic Dis. 2011;11:1447–51.
Špitalská E, Štefanidesová K, Kocianová E, Boldiš V. Rickettsia slovaca and Rickettsia raoultii in Dermacentor marginatus and Dermacentor reticulatus ticks from Slovak Republic. Exp Appl Acarol. 2012;57:189–97.
Švehlová A, Berthová L, Sallay B, Boldiš V, Sparagano OA, Špitalská E. Sympatric occurrence of Ixodes ricinus, Dermacentor reticulatus and Haemaphysalis concinna ticks and Rickettsia and Babesia species in Slovakia. Ticks Tick-borne Dis. 2014;5:600–5.
Minichová L, Hamšíková Z, Mahríková L, Slovák M, Kocianová E, Kazimírová M, et al. Molecular evidence of Rickettsia spp. in ixodid ticks and rodents in suburban, natural and rural habitats in Slovakia. Parasit Vectors. 2017;10:158.
Špitalská E, Sparagano O, Stanko M, Schwarzová K, Špitalský Z, Škultéty Ľ, et al. Diversity of Coxiella-like and Francisella-like endosymbionts, and Rickettsia spp., Coxiella burnetii as pathogens in the tick populations of Slovakia. Central Europe. Ticks Tick Borne Dis. 2018;9:1207–11.
Lapin M, Faško P, Melo M, Šťastný P, Tomlain J. Climatic regions. In: Miklos L, Hrnčiarová T, editors. Landscape atlas of the Slovak Republic. Bratislava and Banská Štiavnica: Ministry of Environment of the Slovak Republic; 2002. p. 95.
Filippova NA. Fauna of the USSR. 4. Chelicerata. Ixodid ticks of the subfamily Ixodinae. Leningrad: Nauka. 1977.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96.
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200.
Westcott SL, Schloss PD. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. MSphere. 2017;2:e00073–117.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335.
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7.
McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610.
Kurilshikov A, Livanova NN, Fomenko NV, Tupikin AE, Rar VA, Kabilov MR, et al. Comparative metagenomic profiling of symbiotic bacterial communities associated with Ixodes persulcatus, Ixodes pavlovskyi and Dermacentor reticulatus ticks. PLoS ONE. 2015;10:e0131413.
Tekin S, Dowd SE, Davinic M, Bursali A, Keskin A. Pyrosequencing based assessment of bacterial diversity in Turkish Rhipicephalus annulatus and Dermacentor marginatus ticks (Acari: Ixodidae). Parasitol Res. 2017;116:1055–61.
Qiu Y, Nakao R, Ohnuma A, Kawamori F, Sugimoto C. Microbial population analysis of the salivary glands of ticks; a possible strategy for the surveillance of bacterial pathogens. PLoS ONE. 2014;9:e103961.
Kwan JY, Griggs R, Chicana B, Miller C, Swei A. Vertical vs. horizontal transmission of the microbiome in a key disease vector, Ixodes pacificus. Mol Ecol. 2017;26:6578–89.
René-Martellet M, Minard G, Massot R, Moro CV, Chabanne L, Mavingui P. Bacterial microbiota associated with Rhipicephalus sanguineus (s.l.) ticks from France, Senegal and Arizona. Parasit Vectors. 2017;10:416.
Estrada-Peña A, Cabezas-Cruz A, Pollet T, Vayssier-Taussat M, Cosson JF. High throughput sequencing and network analysis disentangle the microbial communities of ticks and hosts within and between ecosystems. Front Cell Infect Microbiol. 2018;8:236.
Bona M, Stanko M. First records of the tick Ixodes frontalis (Panzer, 1795) (Acari, Ixodidae) in Slovakia. Ticks Tick Borne Dis. 2013;4:478–81.
SmetanovÁ K, SchwarzovÁ K, KocianovÁ E. Detection of Anaplasma phagocytophilum, Coxiella burnetii, Rickettsia spp., and Borrelia burgdorferi s.l. in ticks, and wild-living animals in western and middle Slovakia. Ann N Y Acad Sci. 2006;1078:312–5.
Zhu YX, Song YL, Hoffmann AA, Jin PY, Huo SM, Hong XY. A change in the bacterial community of spider mites decreases fecundity on multiple host plants. MicrobiologyOpen. 2018;8:e743.
Sperling JL, Silva-Brandao KL, Brandao MM, Lloyd VK, Dang S, Davis CS, et al. Comparison of bacterial 16S rRNA variable regions for microbiome surveys of ticks. Ticks Tick Borne Dis. 2017;8:453–61.
Lalzar I, Harrus S, Mumcuoglu KY, Gottlieb Y. Composition and seasonal variation of Rhipicephalus turanicus and Rhipicephalus sanguineus bacterial communities. Appl Environ Microbiol. 2012;78:4110–6.
Williams-Newkirk AJ, Rowe LA, Mixson-Hayden TR, Dasch GA. Characterization of the bacterial communities of life stages of free-living lone star ticks (Amblyomma americanum). PLoS One. 2014;9:e102130.
Moreno CX, Moy F, Daniels TJ, Godfrey HP, Cabello FC. Molecular analysis of microbial communities identified in different developmental stages of Ixodes scapularis ticks from Westchester and Dutchess counties, New York. Environ Microbiol. 2006;8:761–72.
Zhang XC, Yang ZN, Lu B, Ma XF, Zhang CX, Xu HJ. The composition and transmission of microbiome in hard tick, Ixodes persulcatus, during blood meal. Ticks Tick Borne Dis. 2014;5:864–70.
Rynkiewicz EC, Hemmerich C, Rusch DB, Fuqua C, Clay K. Concordance of bacterial communities of two tick species and blood of their shared rodent host. Mol Ecol. 2015;24:2566–79.
Hernández-jarguín A, Díazsánchez S, Villar M, De JLF. Integrated metatranscriptomics and metaproteomics for the characterization of bacterial microbiota in unfed Ixodes ricinus. Ticks Tick Borne Dis. 2018;9:1241–51.
Díaz-Sánchez S, Hernández-Jarguín A, Torina A, de Mera IGF, Blanda V, Caracappa S, et al. Characterization of the bacterial microbiota in wild-caught Ixodes ventalloi. Ticks Tick Borne Dis. 2019;10:336–43.
Kostanjšek R, Marolt TP. Pathogenesis, tissue distribution and host response to Rhabdochlamydia porcellionis, infection in rough woodlouse Porcellio scaber. J Invertebr Pathol. 2015;125:56–67.
Corsaro D, Thomas V, Goy G, Venditti D, Radek R, Greub G. ‘Candidatus Rhabdochlamydia crassificans’, an intracellular bacterial pathogen of the cockroach Blatta orientalis (Insecta: Blattodea). Syst Appl Microbiol. 2007;30:221–8.
Vanthournout B, Hendrickx F. Endosymbiont dominated bacterial communities in a dwarf spider. PLoS ONE. 2015;10:e0117297.
Croxatto A, Rieille N, Kernif T, Bitam I, Aeby S, Péter O, et al. Presence of Chlamydiales DNA in ticks and fleas suggests that ticks are carriers of Chlamydiae. Ticks Tick-borne Dis. 2014;5:359–65.
Pilloux L, Aeby S, Gaümann R, Burri C, Beuret C, Greub G. The high prevalence and diversity of Chlamydiales DNA within Ixodes ricinus ticks suggest a role for ticks as reservoirs and vectors of Chlamydia-related bacteria. Appl Environ Microbiol. 2015;81:8177–82.
Facco F, Grazi G, Bonassi S, Magnani M, Di Pietro P. Chlamydial and rickettsial transmission through tick bite in children. Lancet. 1992;339:992–3.
Ahantarig A, Trinachartvanit W, Baimai V, Grubhoffer L. Hard ticks and their bacterial endosymbionts (or would be pathogens). Folia Microbiol. 2013;58:419–28.
Fernández-Soto P, Pérez-Sánchez R, Encinas-Grandes A, Sanz RÁ. Rickettsia slovaca in Dermacentor ticks found on humans in Spain. Eur J Clin Microbiol Infect Dis. 2006;25:129.
Parola P, Rovery C, Rolain JM, Brouqui P, Davoust B, Raoult D. Rickettsia slovaca and R. raoultii in tick-borne rickettsioses. Emerg Infect Dis. 2009;15:1105.
Zhang YK, Liu JZ. Maternally inherited symbiotic bacteria in ticks: incidence and biological importance. Syst Appl Acarol. 2019;24:158–80.
Jiggins FM, Hurst GDD, Jiggins CD, vd Schulenburg JHG, Majerus MEN. The butterfly Danaus chrysippus is infected by a male-killing Spiroplasma bacterium. Parasitology. 2000;120:439–46.
Ferree PM, Avery A, Azpurua J, Wilkes T, Werren JH. A bacterium targets maternally inherited centrosomes to kill males in Nasonia. Curr Biol. 2008;18:1409–14.
Acknowledgements
We thank Professor Xiao-Long Yang and Dr Tian-Tian Zhang of Hebei Normal University for their valuable comments on the earlier version of this manuscript.
Funding
This study was supported by Scientific and Technological Cooperation Project of China and Slovakia (7-12; APVV-SK-CN-2015-0010), Advanced Talents of Hebei Normal University (L2018B15), Natural Science Foundation of Hebei province (C2018205211) and National Natural Science Foundation of China (31802008), Project of the Slovak Scientific Grant Agency VEGA (no. 2/0126/16).
Author information
Authors and Affiliations
Contributions
YKZ, JZL and ZJY developed and wrote the manuscript. DW, VB and PB provided critical reviews and suggestions for the content. JZL, VB and PB collected the tick samples. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: Table S1.
Analysis of the obtained MiSeq sequencing reads per sample.
Additional file 2: Figure S1.
Rarefaction curves for Shannonʼs index at OTU level.
Additional file 3: Table S2.
Analysis of taxonomy and abundance of the obtained OTUs.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, YK., Yu, ZJ., Wang, D. et al. The bacterial microbiome of field-collected Dermacentor marginatus and Dermacentor reticulatus from Slovakia. Parasites Vectors 12, 325 (2019). https://doi.org/10.1186/s13071-019-3582-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3582-9