Abstract
Background
Enterocytozoon bieneusi is an obligate, intracellular fungus and is commonly reported in humans and animals. To date, there have been no reports of E. bieneusi infections in Bactrian camels (Camelus bactrianus). The present study was conducted to understand the occurrence and molecular characteristics of E. bieneusi in Bactrian camels in China.
Results
Of 407 individual Bactrian camel fecal specimens, 30.0% (122) were E. bieneusi-positive by nested polymerase chain reaction (PCR) based on internal transcriber spacer (ITS) sequence analysis. A total of 14 distinct E. bieneusi ITS genotypes were obtained: eight known genotypes (genotype EbpC, EbpA, Henan-IV, BEB6, CM8, CHG16, O and WL17), and six novel genotypes (named CAM1 to CAM6). Genotype CAM1 (59.0%, 72/122) was the most predominant genotype in Bactrian camels in Xinjiang, and genotype EbpC (18.9%, 23/122) was the second-most predominant genotype. Phylogenetic analysis revealed that six known genotypes (EbpC, EbpA, WL17, Henan-IV, CM8 and O) and three novel genotypes (CAM3, CAM5 and CAM6) fell into the human-pathogenic group 1. Two known genotypes (CHG16 and BEB6) fell into the cattle host-specific group 2. The novel genotypes CAM1, CAM 2 and CAM4 cluster into group 8.
Conclusions
To our knowledge, this is the first report of E. bieneusi in Bactrian camels. The host-specific genotype CAM1 was the predominant genotype, which plays a negligible role in the zoonotic transmission of E. bieneusi. However, the second-most predominant genotype, EbpC, has greater zoonotic potential.
Similar content being viewed by others
Background
Microsporidia are a diverse group of emerging obligate intracellular eukaryotic fungi and there are approximately 1300 microsporidian species in 160 genera [1]. To date, there are at least 14 microsporidian species reported to be infectious to humans [2]. Enterocytozoon bieneusi is the most frequently detected species in humans [3], as well as in domestic animals and wildlife [4], and even in environmental water samples [5].
More than 200 E. bieneusi genotypes have been identified in humans and animals by polymerase chain reaction (PCR) based on ribosomal internal transcribed spacer (ITS) gene sequence analysis [2, 6]. Molecular phylogenetic analysis has shown that all E. bieneusi ITS genotypes are clustered into nine large groups, including the potentially zoonotic group 1, and some host-specific groups (Group 2 to Group 9) [7].
The Bactrian camel (Camelus bactrianus) was the major means of transportation on the ancient Silk Road. Today, the population of Bactrian camels in China has been estimated at 242,000, most of which are domesticated in desert and semi-desert areas of northwestern China and play an important role in the livelihood of pastoralists through providing milk and meat [8]. There are some reports of intestinal pathogen infections in camels and Bactrian camels in the Middle East countries and China, such as Eimeria spp. and Cryptosporidium spp. [9, 10]. However, E. bieneusi infection has not been previously reported in Bactrian camels.
This study was undertaken to better understand the prevalence of E. bieneusi in Bactrian camels and assess the host specificity of E. bieneusi infections in Bactrian camels in China.
Methods
Specimen collection
A total of 407 individual fresh fecal specimens from Bactrian camels were collected from 18 different grazing Bactrian camel groups in 11 collection sites of Xinjiang Uygur Autonomous Region (hereinafter referred to as Xinjiang) of northwestern China. Only one specimen was collected per animal. These specimens were collected during August and September of 2013 and from July 2016 to July 2017 (Table 1). The grazing Bactrian camel groups were kept outdoors and shared pastures with cattle, sheep, goats and wild animals, and each group had approximately 30–300 animals.
After animal defecation, about 50–100 g of each fresh specimen was collected immediately from the ground using sterile gloves. Each specimen was collected in a plastic container and marked with the specimen number and site. The specimens were transported to the laboratory and stored in 2.5% (w/v) potassium dichromate solution at 4 °C before DNA extraction.
DNA extraction and PCR amplification
Approximately 200 mg of each fecal specimen was washed at least three times with distilled water by centrifugation at 5000× g for 5 min to remove the potassium dichromate. DNA was extracted using the E.Z.N.A.R® Stool DNA Kit (Omega Biotek Inc., Norcross, GA, USA) according to the manufacturer’s instructions. For E. bieneusi screening, nested PCR assays were used to amplify an rRNA gene fragment containing the entire internal transcriber spacer (ITS) [6]. Each specimen was analyzed in duplicate using positive and negative controls. The secondary PCR products were examined by electrophoresis in a 1.5% agarose gel and visualized after staining with GelRed™ (Biotium Inc., Hayward, CA, USA).
Sequencing and phylogenetic analysis
The positive secondary PCR amplicons were sent to a commercial company (GENEWIZ, Suzhou, China) for sequencing. The sequence accuracy was confirmed with bidirectional sequencing, and the sequences obtained were aligned with reference sequences downloaded from GenBank to determine the genotypes, using the program ClustalX 2.0 (http://www.clustal.org/).
The genotypes of E. bieneusi isolated in this study were compared with known E. bieneusi ITS genotypes with a neighbor-joining analysis in the Mega 5 program [6]. A bootstrap analysis was used to assess the robustness of the clusters using 1000 replicates. The established nomenclature system was used in naming the E. bieneusi ITS genotypes [11].
Nucleotide sequence accession numbers
The nucleotide sequences reported in this paper have been submitted to the GenBank database at the National Center for Biotechnology Information under the accession numbers: MG602791-MG602796.
Statistical analysis
Chi-square test was used to compare the prevalence of E. bieneusi infections and predominant genotypes distributions. Differences were considered significant at P < 0.05.
Results and discussion
Of all 407 individual Bactrian camel fecal specimens, 30.0% (122) were E. bieneusi-positive based on the ITS sequence analysis. The majority of the grazing Bactrian camel groups, 14 out of 18 (77.8%), were positive for E. bieneusi. Among them, Urumqi-1 had the highest infection rate (84.6%, 11/13) (χ2 = 67.728, df = 17, P < 0.001); the other infection rates ranged from 16.7–52.9% (Table 1).
To the best of our knowledge, this is the first report of E. bieneusi in Bactrian camels, and the pathogen is widespread in Xinjiang, northwestern China. In China, the average prevalence of E. bieneusi in animals ranges from 0.9% (4/426) in rabbits [12] to 45.6% (426/934) in pigs [13]. However, E. bieneusi infection has only been reported in some animals in northwestern China (Table 2), the average prevalence ranging from 1.1% (4/353) in white yaks [14] to 47.8% (22/46) in sheep [2]. In Xinjiang, only dairy calves [15] and grazing horses [16] have been previously reported to have E. bieneusi infections, with a prevalence of 16.5% (85/514) and 30.9% (81/262), respectively. The high prevalence in Bactrian camels found in this study may be the result of free feeding and drinking water, and mixed feeding with cattle, sheep, goats and other animals in the same pastures, and with the poor veterinary service.
A total of 14 distinct E. bieneusi ITS genotypes were obtained from 122 positive specimens from Bactrian camels. Among them, eight were known genotypes (EbpC, EbpA, Henan-IV, BEB6, CM8, CHG16, O and WL17), and six were novel genotypes (named CAM1 to CAM6). The sequences of the novel genotypes, CAM1 to CAM6, consisted of insertions, deletions, and substitutions compared with known genotypes. In the present study, the novel genotype CAM1 was the most prevalent (59.0%, 72/122) and was significantly predominant (χ2 = 589.836, df = 13, P < 0.001) in Bactrian camels in Xinjiang. The other novel genotypes CAM2 (n = 8), CAM4 (n = 5), CAM3 (n = 1), CAM5 (n = 1) and CAM6 (n = 1) were also identified. The genotype EbpC was the predominant identified of the known genotypes (18.9%, 23/122) in Bactrian camels in Xinjiang, followed by genotype EbpA (n = 5). However, the other known genotypes Henan-IV, BEB6, CM8, CHG16, O and WL17, were identified in only one specimen each, though they have been commonly reported in many other types of animals.
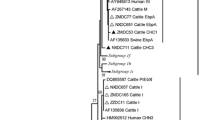
The phylogenetic analysis of the ITS genotypes revealed the following clusters: Group 1, Group 2 and Group 8. The six known genotypes (EbpC, EbpA, WL17, Henan-IV, CM8 and O) and three novel genotypes (CAM3, CAM5 and CAM6) identified in this study fell into the human-pathogenic Group 1 (Fig. 1), which is the genotype of major zoonotic potential suggesting that the Bactrian camels play a potential role in E. bieneusi transmission to humans [11]. In contrast, the two known genotypes CHG16 and BEB6 fell into the cattle host-specific Group 2. The three novel genotypes CAM1, CAM 2 and CAM4 clustered into Group 8 (69.7%, 85/122) (Fig. 1), suggesting that the host-specific genotype CAM1 in Bactrian camels exhibits less zoonotic potential compared to the genotypes clustered into the human-pathogenic group.
Phylogenetic relationships of the E. bieneusi genotypes identified in this study and other reported genotypes. The phylogeny was inferred with a neighbor-joining analysis of the internal transcribed spacer (ITS) sequences based on distances calculated with the Kimura two-parameter model. Bootstrap values > 50% from 1000 replicates are shown at the nodes. The genotypes detected in this study are shown with triangles; previously known genotypes observed in this study are marked with open triangles and the new genotypes are indicated by filled triangles
In previous studies, E. bieneusi genotype EbpC and EbpA were reported in humans and various animals and were also the predominant genotypes in the reports of humans and pigs in China [13, 17]. Genotypes EbpC and EbpA were the most common E. bieneusi genotypes in grazing horses in Xinjiang [16], and genotype EbpC was also identified in dairy calves in Xinjiang [15]. Similarly, the E. bieneusi zoonotic genotypes EbpC and EbpA were identified in Bactrian camels in the present study. However, there were no published reports of genotypes EbpC and EbpA in animals in Gansu, Ningxia, Qinghai and Shaanxi, northwestern China (Table 2). Further investigations of the epidemiology and host specificity of E. bieneusi in humans and other animals in Xinjiang might be informative.
Conclusions
The present study demonstrated a widespread occurrence of E. bieneusi in Bactrian camels in Xinjiang, China. The host-specific genotype, CAM1, was the most predominant genotype, which plays a negligible role in the zoonotic transmission of E. bieneusi. The second-most predominant genotype, EbpC, in addition to other genotypes of zoonotic potential, was also commonly identified in Bactrian camels in this study. Bactrian camels could serve as a vector for E. bieneusi transmission to humans and other animals, and vice versa.
Abbreviations
- ITS:
-
Ribosomal internal transcribed spacer
- PCR:
-
Polymerase chain reaction
References
Keeling P. Five questions about microsporidia. PLoS Pathog. 2009;5(9):e1000489.
Shi K, Li M, Wang X, Li J, Karim MR, Wang R, et al. Molecular survey of Enterocytozoon bieneusi in sheep and goats in China. Parasit Vectors. 2016;9:23.
Liu H, Jiang Z, Yuan Z, Yin J, Wang Z, Yu B, et al. Infection by and genotype characteristics of Enterocytozoon bieneusi in HIV/AIDS patients from Guangxi Zhuang Autonomous Region, China. BMC Infect Dis. 2017;17(1):684.
Santín M, Fayer R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci. 2011;90:363–71.
Guo Y, Alderisio KA, Yang W, Cama V, Feng Y, Xiao L. Host specificity and source of Enterocytozoon bieneusi genotypes in a drinking source watershed. Appl Environ Microbiol. 2014;80(1):218–25.
Li J, Luo N, Wang C, Qi M, Cao J, Cui Z, et al. Occurrence, molecular characterization and predominant genotypes of Enterocytozoon bieneusi in dairy cattle in Henan and Ningxia, China. Parasit Vectors. 2016;9:142.
Karim MR, Dong H, Li T, Yu F, Li D, Zhang L, et al. Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: high genetic diversity and zoonotic significance. PLoS One. 2015;10(2):e0117991.
China National Commission of Animal Genetic Resources. Animal genetic resources in China: horses, donkeys, camels. Beijing: China Agricultural Press; 2011. p. 2–6. (In Chinese)
Liu X, Zhou X, Zhong Z, Deng J, Chen W, Cao S, et al. Multilocus genotype and subtype analysis of Cryptosporidium andersoni derived from a Bactrian camel (Camelus bactrianus) in China. Parasitol Res. 2014;113(6):2129–36.
Sazmand A, Joachim A. Parasitic diseases of camels in Iran (1931–2017) - a literature review. Parasite. 2017;24:21.
Thellier M, Breton J. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite. 2008;15(3):349–58.
Zhang XX, Jing J, Cai YN, Wang CF, Peng X, Yang GL, et al. Molecular characterization of Enterocytozoon bieneusi in domestic rabbits (Oryctolagus cuniculus) in northeastern China. Korean J Parasitol. 2017;54(1):81–5.
Li W, Diao R, Yang J, Xiao L, Lu Y, Li Y, et al. High diversity of human-pathogenic Enterocytozoon bieneusi genotypes in swine in northeast China. Parasitol Res. 2014;113(3):1147–53.
Ma JG, Zhang NZ, Hou JL, Zou Y, Hu GX, Zhu XQ, et al. Detection of Enterocytozoon bieneusi in White Yaks in Gansu Province, China. Biomed Res Int. 2017;2017:5790181.
Qi M, Jing B, Jian FC, Wang RJ, Zhang SM, Wang HY, et al. Dominance of Enterocytozoon bieneusi genotype J in dairy calves in Xinjiang, northwest China. Parasitol Int. 2017;66(1):960–3.
Qi M, Wang RJ, Wang HY, Jian FC, Li JQ, Zhao JF, et al. Enterocytozoon bieneusi genotypes in grazing horses in China and their zoonotic transmission potential. J Eukaryot Microbiol. 2016;63(5):591–7.
Wang L, Zhang HW, Zhao XD, Zhang LX, Zhang GQ, Guo MJ, et al. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol. 2013;51(2):557–63.
Ma J, Cai J, Ma J, Feng Y, Xiao L. Enterocytozoon bieneusi genotypes in yaks (Bos grunniens) and their public health potential. J Eukaryot Microbiol. 2015;62(1):21–5.
Du SZ, Zhao GH, Shao JF, Fang YQ, Tian GR, Zhang LX, et al. Cryptosporidium spp., Giardia intestinalis, and Enterocytozoon bieneusi in captive non-human primates in Qinling Mountains. Korean J Parasitol. 2015;53(4):395–402.
Wang X, Wang R, Ren G, Yu Z, Zhang L, Zhang S, et al. Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi Province, northwestern China. Parasitol Res. 2016;115(3):1355–61.
Zhao GH, Du SZ, Wang HB, Hu XF, Deng MJ, Yu SK, et al. First report of zoonotic Cryptosporidium spp., Giardia intestinalis and Enterocytozoon bieneusi in golden takins (Budorcas taxicolor bedfordi). Infect Genet Evol. 2015;34:394–401.
Peng XQ, Tian GR, Ren GJ, Yu ZQ, Lok JB, Zhang LX, et al. Infection rate of Giardia duodenalis, Cryptosporidium spp. and Enterocytozoon bieneusi in cashmere, dairy and meat goats in China. Infect Genet Evol. 2016;41:26–31.
Acknowledgments
We thank Liwen Bianji, Edanz Editing China (http://www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This study was partly supported by the National Key Research and Development Program of China (2017YFD0501305), and the Key Laboratory of Tarim Animal Husbandry, Science, and Technology, Xinjiang Production & Construction Group Open Project Fund (HS201714Z), and the President Foundation of Tarim University (TDZKJC201601, TDZKCX201701).
Availability of data and materials
The data sets supporting the conclusions of this article are included within the article. The newly generated sequences were deposited in the GenBank database under the accession numbers MG602791-MG602796.
Author information
Authors and Affiliations
Contributions
LXZ and MQ designed the study. BJ, AYZ and ZLW participated in specimen collection and analysis. JQL, BJ, MQ and ZHC contributed to data analysis. LXZ, JQL and MQ wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was conducted in accordance with the Chinese Laboratory Animal Administration Act (1988). The study protocol was reviewed and approved by the Research Ethics Committee of Tarim University. Appropriate permission was obtained from the owner before the collection of fecal specimens.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Qi, M., Li, J., Zhao, A. et al. Host specificity of Enterocytozoon bieneusi genotypes in Bactrian camels (Camelus bactrianus) in China. Parasites Vectors 11, 219 (2018). https://doi.org/10.1186/s13071-018-2793-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-018-2793-9