Abstract
Background
Human babesiosis is an infectious disease that is epidemic in various regions all over the world. The predominant causative pathogen of this disease is the intra-erythrocytic parasite Babesia microti. The thioredoxin system is one of the major weapons that is used in the resistance to the reactive oxygen species (ROS) and reactive nitrogen species (RNS) produced by host immune system. In other intra-erythrocytic apicomplexans like the malaria parasite Plasmodium falciparum, anti-oxidative proteins are promising targets for the development of anti-parasitic drugs. However, to date, the sequences and biological properties of thioredoxins and thioredoxin-like molecules of B. microti remain unknown. Understanding the molecular characterization and function of B. microti thioredoxins may help to develop anti-Babesia drugs and controlling babesiosis.
Methods
The full-length B. microti thioredoxin 2 (BmTrx2) gene was obtained using a rapid amplification of cDNA ends (RACE) method, and the deduced BmTrx2 amino acid sequence was analyzed using regular bioinformatics tools. Recombinant BmTrx2 protein was expressed in vitro and purified using His-tag protein affinity chromatography resins. Reverse transcription PCR, quantitative real-time PCR and Western blot were employed to detect the expression and native proteins of BmTrx2. Indirect immunofluorescence assay was used to localize BmTrx2 in B. microti. Bovine insulin reduction assays were used to determine the enzyme activity of the purified recombinant BmTrx2 protein.
Results
The full-length BmTrx2 was 564 bp with a 408 bp open reading frame encoding a protein of 135 amino acids. The predicted molecular weight of the protein was 15.5 kDa. A conserved thioredoxin-like family domain was found in BmTrx2. The expression of BmTrx2 was upregulated on both the third and eighth day post-infection in mice, whereas expression was downregulated during the beginning and later stages. The results of Western blot analysis showed the native BmTrx2 in parasite lysates could be detected by mouse anti-BmTrx2 serum and that the recombinant BmTrx2 protein could be recognized by serum of B. microti-infected mice. Immunofluorescence microscopy showed that BmTrx2 localized in the cell cytoplasm of B. microti merozoites in B. microti-infected red blood cells. The results of bovine insulin reduction assay indicated the purified recombinant BmTrx2 protein possesses antioxidant enzyme activity. Dihydroartemisinin and quinine, known anti-malaria drugs, and clindamycin, a known anti-babesiosis drug, induced significantly higher upregulation of BmTrx2 mRNA.
Conclusions
Our results indicate that BmTrx2 is a functional enzyme with antioxidant activity and may be involved in the response of B. microti to anti-parasite drugs.
Similar content being viewed by others
Background
Babesia microti is a tick-borne protozoan parasite belonging to the phylum Apicomplexa that invades and replicates in the erythrocytes of animals, including humans, causing zoonotic babesiosis [1]. There are over 100 different species of the genus Babesia, including B. microti, B. bovis, B. divergens and B. bigemina, that range in size between 1 and 5 μm [2]. Since the first report of human infection with Babesia in 1957, thousands of human infection cases have been reported around the world, making babesiosis an important emerging human infectious disease [3]. The most common symptoms of severe infection Babesia are hemolytic anaemia and jaundice. Severe symptoms occur in susceptible patients, such as those who are asplenic or immunosuppressed. In 2011, the first two humans infected with B. divergens in China were reported in Shandong Province [4]. Given its prevalence and severity, and because there are currently no effective drugs to treat babesiosis, the need for the identification of new drug targets is urgent.
Reactive oxygen species (ROS) and reactive nitrogen species (RNS) in the high oxygen environment of mammalian hosts’ red blood cells (RBCs) are toxic to Babesia trophozoites and merozoites. Therefore, to survive in the host, the parasites must protect themselves from elimination by ROS [5]. However, because these parasites lack antioxidant enzymes, such as catalase and glutathione peroxidase, the thioredoxin system plays a significant role in their resistance to host oxidative stress [6, 7]. The thioredoxin system consists of three major components, thioredoxin (Trx), thioredoxin reductase (TrxR) and NADPH, of which Trx plays a central role in redox regulation and antioxidation [8]. Other apicomplexan parasites, such as Plasmodium falciparum, are sensitive to disturbances in the antioxidant system, making it a potential target for novel antiprotozoal drugs [7]. Previous studies showed that five Trx genes had been found in P. falciparum. Among them, thioredoxin 1–3 can be reduced by PfTrxR; however thioredoxin-like protein 1 and 2 cannot [9]. Trx has already been studied as a potential drug target [7]. In Babesia, other antioxidant molecules have been reported, including superoxide dismutase, catalase, and glutathione reductase [10, 11]. However, no research on the thioredoxins of Babesia has been reported. In the present study, the B. microti Trx2 (BmTrx2) gene was cloned and characterized, and its role in response to antiprotozoal drugs was tested.
Methods
Parasites and animals
The B. microti strain ATCC® PRA-99TM was obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in our laboratory by serial passage in BALB/C mice (SLAC, Shanghai, China) using the method described previously [12].
Transcriptome sequencing of B. microti
The B. microti total RNA sequencing using Illumina Hiseq 4000 was performed by BGI (The Beijing Genomics Institute, Shenzhen, China) as described previously [12].
Molecular cloning and sequence analysis of BmTrx2
BmTrx2 was selected from the cDNA library constructed previously, for further study. Total RNA was extracted from erythrocytic stage B. microti using a routine method as described elsewhere [12]. For the synthesis of complementary DNAs (cDNAs), reverse transcription PCR (RT-PCR) was performed using PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China), according to the instructions. The partial coding sequence of BmTrx2 has been deposited in the GenBank database (reference sequence: XM_021482575.1) [13]. However, the complete open reading frame (ORF) of BmTrx2 was not available. The full-length BmTrx2 was generated by rapid amplification of cDNA ends (RACE) using SMARTer RACE cDNA amplification kit (Clontech, Mountain view, USA) according to the manufacturer’s instructions. The full-length BmTrx2 ORF was amplified using primers designed based on the sequence which was generated by RACE. The primer sets used in this study are shown in Table 1. The amplified PCR products were subsequently purified and subcloned into the pMD-19 T vector (TaKaRa) and further confirmed by sequencing, which was performed at Genewiz (Genewiz Inc., Suzhou, China). Finally, the identified sequence was submitted to the NCBI database.
The software Genetyx (Software Development Co., Ltd., Tokyo, Japan) was used to analyze the nucleotide and the deduced amino acid sequences. As for sequence homology, Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) was employed for the assessment. The phylogenetic tree was constructed using ClustalW alignment and neighbour-joining method of the software Mega 6.06.
In vitro expression and purification of recombinant BmTrx2 protein
The ORF of BmTrx2 was amplified by polymerase chain reaction (PCR) using the previously described primers and directionally cloned into the pET-30a(+) vector (Novagen, Madison, USA). The recombinant plasmid pET-30a(+)-BmTrx2 was verified by sequencing and then transferred into Escherichia coli BL21 (DE3) (Novagen) for expression of a six His-tagged recombinant protein. Positive clones were cultured in LB media and induced with the isopropyl β-D-thiogalactoside (IPTG) (1μg/ml) at 16 °C for 12 h (optical density at 600 nm OD600 = 0.6).
Recombinant BmTrx2 protein was purified using Ni-NTA His·Bind Resin (Millipore, Burlington, USA), following instructions of the manufacturer. The purity of the purified protein was confirmed by SDS-PAGE on 12% electrophoretic gels, and the concentration was measured using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, USA). Pierce High Capacity Endotoxin Removal Spin Columns (Thermo Fisher Scientific) was utilized to remove the endotoxin from protein samples used for in vivo trials. The purified protein was aliquoted and stored at -80 °C for further use.
Immune serum production
A polyclonal antibody against rBmTrx2 was obtained using a method described previously [14] except the purified rBmTrx2 were used in our study. The serum was collected 2 weeks after the third immunization. Specific anti-BmTrx2 antibody titers were assessed by enzyme-linked immunosorbent assay (ELISA) [15].
Anti-B. microti serum was collected from BALB/C mice 21 days post-infection with 1 × 108 B. microti-infected RBCs (iRBCs) with a parasitemia of 30%. The immune serum was aliquoted and stored at -80 °C until further use.
Western blot analysis of the native and recombinant BmTrx2 proteins
The Western blot analysis was performed using a regular method described previously [14]. Briefly, samples from five batches of somatic extract of iRBCs (5, 6, 7 and 8 days post-infection and non-infected erythrocytes as a negative control) and the purified rBmTrx2 were fractioned by SDS-PAGE. Gel-separated proteins were then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). Then, primary antibodies, mouse anti-rBmTrx2 sera (dilutions 1:200) or mouse anti-B. microti sera (dilutions 1:200) were applied, respectively. Finally, a secondary antibody HRP-conjugated goat-anti-mouse IgG (dilutions 1:2000) (Sigma-Aldrich, St. Louis, USA) was added and the protein-antibody complex was developed using an Enhanced DAB kit (Tiangen Biotech, Beijing, China) following to the manufacturer’s instructions.
Relative expression analysis of BmTrx2 post-infection
The determination of BmTrx3 expression post-infection was performed as described previously [16] except the 18S ribosomal RNA of B. microti (Bm18S) (GenBank: XM_021481625.1) [13, 17] was used as an internal control in this study. Briefly, Mice were injected with 1 × 108 iRBCs, and the blood was collected from 1 to 10 days post-injection. Total RNAs were extracted from iRBCs at different post-infection time points for qRT-PCR analysis. The specific primers used to quantify BmTrx2 and Bm18S are shown in Table 1. Also, the parasitemia was calculated at each day post-infection.
Expression of BmTrx2 after treatment with antiprotozoal drugs
In this study, a B. microti iRBCs short-term in vitro culture system was established. Briefly, B. microti-infected mice blood (30% parasitemia) was collected by cardiac puncture after anaesthesia. iRBCs were washed four times with sterile PBS. For drug screening, a 12-well flat-bottom plate (Nunc cell culture, USA) was used. A total of 2 × 107 B. microti iRBCs were cultured in basic media (RPMI 1640 containing 40% fetal bovine serum [FBS], 25 mM HEPES) at 37 °C in an atmosphere of 95% air and 5% CO2 [18, 19].
To assess the effect of drugs on BmTrx2 gene expression, the short-term in vitro growth of iRBCs against four antiprotozoal drugs (dihydroartemisinin, quinine, chloroquine and clindamycin) (Sigma-Aldrich) was tested. Dimethylsulfoxide (DMSO) was used to dissolve dihydroartemisinin, quinine, and clindamycin, whereas PBS was utilized to dilute chloroquine. iRBCs were treated with different concentrations (20, 50, or 100 μM) of dihydroartemisinin, quinine, chloroquine, and clindamycin for 24 h, and controls were treated with DMSO or PBS. iRBCs were processed as described above and relative BmTrx2 transcript levels were assessed.
Indirect immunofluorescence assay
The indirect immunofluorescence assay was conducted using a conventional method described elsewhere [16] with slight modifications. Serum of mice hyper-immunized against rBmTrx2 was generated by immunizing animals three times. B. microti iRBCs smears were fixed with methanol and acetone (1:1, v/v) and permeabilized with 0.1% TritonX-100. After washing, primary antibody, mouse anti-BmTrx2 serum diluted 1:500 in 3% BSA was applied. The secondary antibody used in this study was Alexa-Fluor®488 conjugated goat anti-mouse IgG (Invitrogen, Carlsbad, USA). Nucleus stain was performed by incubating the smears with 2 μg/ml Hoechst 33342 (Thermo Fisher). Finally, the slides were examined using a confocal laser-scanning microscope (Zeiss LSM 880, Oberkochen, Germany) after mounting with Fluoromount Aqueous Mounting Media (Sigma-Aldrich). The parameters in all the experiment and control groups were set at the same values.
Enzyme activity assays
The evaluation of rBmTrx2 enzyme activity was conducted using a bovine insulin reduction assay described previously [20]. Briefly, 16 mM bovine insulin (Sigma-Aldrich), 10 μM purified rBmTrx2, 100 nM rat thioredoxin reductase (rat TrxR), 100 μM NADPH and 2 mM EDTA in 50 mM Tris-HCI (pH 6.5) were mixed in a reaction system. Thioredoxin activity was determined by measuring the decrease in absorbance at 340 nm. His fusion protein (10 μM) and His-tagged E. coli thioredoxin (10 μM) were set as controls. All reactions were performed in triplicate for each enzyme.
Data analysis
A GraphPad PRISM 5 software (GraphPad Software Inc., CA, USA) was employed for the statistical analysis. The mean ± standard error (SEM) of each group was calculated. The differences between groups were determined using two-tailed t-tests. P < 0.05 was considered significant and P < 0.01 was considered highly significant.
Results
Cloning of the BmTrx2 gene
Transcriptome sequencing of B. microti revealed 3024 genes obtained from the transcriptome assembly. The partial coding sequences of a thioredoxin reductase and six thioredoxins were present. By multiple alignments using BLAST, these identified genes were named as B. microti thioredoxin reductase 1 [20] and Trx 1–6 (GenBank: KX758048–KX758053) based on the similarity to the thioredoxin genes found in the open database of apicomplexans including Babesia, Plasmodium and Theileria. For the present research, BmTrx2 (KX758049.1) (https://www.ncbi.nlm.nih.gov/nuccore/1235975689) was chosen for further study.
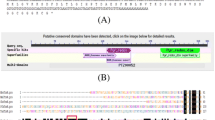
Results of RACE and sequencing showed that the full-length BmTrx2 gene was 564 bp with an ORF of 408 bp encoding a protein of 135 amino acids. Bioinformatics predicted no signal peptides within the BmTrx2 protein. The molecular weight of the predicted protein was 15.5 kDa, and the isoelectric point was 8.77. The results of multiple alignments revealed that the amino acids sequence of the BmTrx2 protein shared 38% similarity with B. bigemina Trx (XP_012765929), 45% similarity with B. bovis Trx (XP_001609888), and 45% similarity with Theileria equi Trx (XP_004828582) (Fig. 1). The results of the phylogenetic analysis revealed that BmTrx2 was most closely related to Theileria (Fig. 2). A thiol-disulfide redox active center (-WCGPC-), which is the typical catalytic domain of Trx, was identified in the amino acid sequence of the BmTrx2 protein (Fig. 1). These results showed that the Trx2 obtained from the mRNA of B. microti merozoites is a typical thioredoxin family protein.
In vitro expression and purification of rBmTrx2
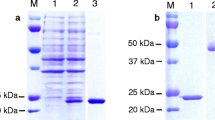
The recombinant BmTrx2 protein was successfully expressed in E. coli as a His-tagged fusion protein and purified using Ni-affinity chromatography. SDS-PAGE results showed that the molecular weight of rBmTrx2 was 15.5 kDa, which was in accordance with the predicted size (Fig. 3). Purified rBmTrx2 were used to obtain mouse anti-BmTrx2 polyclonal serum after 3 immunizations.
Serological detection of native and recombinant BmTrx2 protein
An antiserum obtained from mice immunized with rBmTrx2 was utilized to detect the native BmTrx2 protein. The results of Western blot assays showed that a specific protein band of approximately 15.5 kDa was detected in the iRBC lysates 5, 6, 7 and 8 days post-infection, whereas no bands were detected in the un-infected homogenates (Fig. 4). The molecular size of the detected bands was in line with the expected size of the natural BmTrx2. No other bands were recognized by the antiserum.
Western blot analysis of the native BmTrx2. Lane M: standard protein molecular weight marker; Lanes 1–4: B. microti infected mouse erythrocyte lysates on 5th, 6th, 7th and 8th day post-infection; Lane 5: uninfected mouse erythrocyte lysate; mouse anti-rBmTrx2 serum was used as primary antibody in this Western blot analysis
Western blot analysis also indicated that rBmTrx2 was identified by serum of mice experimentally infected with B. microti, whereas the serum of non-infected mice did not detect rBmTrx2 (Fig. 5). These results suggest that BmTrx2 protein has high immunogenicity and can induce hosts immune response.
Western blot analysis of the recombinant BmTrx2 protein. Lane M: standard protein molecular weight marker; Lane 1: purified rBmTrx2 probed by serum from mice infected by B. microti on 21st d; Lane 2: purified rBmTrx2 probed by uninfected mouse serum; the samples and markers were transferred into the same membrane, after that each lane was separated
Relative expression analysis of BmTrx2 post-infection
To investigate the expression profile of BmTrx2 over time, total RNA of iRBCs from different days post-infection was assessed by qRT-PCR analysis. BmTrx2 expression peaked 3 days post-infection and declined suddenly until 7 days post-infection. Eight days post-infection, the relative expression of BmTrx2 peaked again and then rapidly decreased to low levels (Fig. 6).
Blood obtained via the caudal vein was used to prepare smears which were stained with Giemsa to assess the advancement of the infection by calculating the ratio of iRBCs. Starting on the first day post-infection, B. microti was detected in iRBCs. The parasitemia ratio increased until 5 days post-infection, after which it started to decline until reaching undetectable levels by day 10 post-infection. No parasites were detected in the uninfected mice throughout the selected time points (Fig. 6).
BmTrx2 response to chloroquine, clindamycin, dihydroartemisinin and quinine
mRNA relative expression levels of BmTrx2 were investigated after subjected to different doses of chloroquine, clindamycin, dihydroartemisinin or quinine for 24 h (Fig. 7). Results showed that BmTrx2 was significantly upregulated after treatment with dihydroartemisinin, clindamycin or quinine compared with the control group, whereas no significant difference was observed after chloroquine treatment (Additional file 1: Table S1). Furthermore, quinine-induced BmTrx2 expression was dose-dependent (Fig. 7). These findings indicate that BmTrx2 might be implicated in the response of B. microti to clindamycin, dihydroartemisinin and quinine.
Immunofluorescence assays
The subcellular location of BmTrx2 in B. microti was determined by indirect immunofluorescence assays. B. microti-infected RBCs (approximately 20% parasitemia) and mouse anti-rBmTrx2 serum were used. As shown in Fig. 8, blue fluorescence represents the nucleus of B. microti cells and, green fluorescence represents BmTrx2 located around the nucleus of B. microti merozoites in iRBCs (Fig. 8a). In contrast, no green fluorescence was detected in the iRBCs incubated with non-immune mouse serum (Fig. 8b). Similarly, only blue fluorescence was detected in iRBCs incubated with only second antibody (Fig. 8c). No fluorescence staining was visualized in control mice RBCs incubated with mouse anti-rBmTrx2 serum (Fig. 8d). These results indicate that BmTrx2 is expressed in the cytoplasm of B. microti merozoites.
Immunofluorescence microscopy of native BmTrx2. Observation of the native BmTrx2 recognized by mouse anti-rBmTrx2 serum in confocal laser micrographs (a) iRBCs incubated with mouse anti-rBmTrx2 serum as primary antibody; (b) iRBCs incubated with normal mouse serum as primary antibody; (c) iRBCs incubated with only secondary antibody; (d) uninfected RBCs incubated with mouse anti-rBmTrx2 serum as primary antibody; Scale-bars: 2 μm
Determination of enzyme activity
Enzyme activity of purified rBmTrx2 was analyzed by bovine insulin reduction assays. His-tag was fused to rBmTrx2 protein, and His-tag polypeptide was used as a control. As shown in Fig. 9, absorbance of the His-tag polypeptide remained stable throughout the assay, suggesting that His-tagged polypeptide has no enzymatic activity. In comparison to the positive control (E. coli Trx and rat TrxR), the results showed that the activity of rBmTrx2 was low for the first 100 s of the assay. The absorbance at 340 nm of NADPH gradually decreased at 100–240 s indicating that rBmTrx2 did possess enzymatic activity. After 240 s, the absorbance remained at low levels.
Discussion
Babesia microti, similar to the malaria parasite P. falciparum, is highly equipped with tools to counter the impact of the oxidative stress when the parasites are exposed during the erythrocytic stages. Because intracellular parasites like these apicomplexan protozoa lack antioxidant enzymes such as catalase and glutathione peroxidase, they rely on the thioredoxin system to resist host oxidative stress [21]. Thioredoxin (Trx), thioredoxin reductase (TrxR) and NADPH are involved in the thioredoxin system, and they are widely present in prokaryotes and eukaryotes. The multifaceted functions played by Trx that indirectly depend on TrxR mainly include the modification of the redox status of protein targets via a Cys-xx-Cys active site motif [8]. In this investigation, an essential component of the B. microti thioredoxin system, BmTrx2 was characterized, and the biochemical properties were identified.
Survival of intraerythrocytic P. falciparum emphasized and confirmed how essential thioredoxin reductase is. Previous studies have proved the essential role of thioredoxin reductase in P. falciparum and also the crucial significance of the thioredoxin redox cycle [22]. Other studies reported that Trxs had been identified as potential drug targets for the development antimalarial drugs [23]. In P. falciparum, three classic thioredoxins (Trx 1–3) and two thioredoxin-like proteins (Tlp1 and Tlp2) have been identified [8]. Similarly, six thioredoxin-associated genes were identified in the transcriptome of the B. microti. Using BLAST, the sequence identity of B. microti Trx2 and B. bigemina Trx (XP_012765929) was 38% showing a low homology. However, a typical thiol-disulfide redox active center -Trp-Cys-Gly-Pro-Cys- (-WCGPC-), the Trx catalytic domain critical for regulation of the formation of redox-active disulfide bonds, was identified in the sequence of BmTrx2. The highly conserved Trp and Asp residues provided a hydrogen bonding so that the active site is stable [24]. Also, other studies reported that the highly conserved domain is essential in the maintenance of the three-dimensional structure of Trx [25]. In our study, sequence analysis revealed that the redox active center of BmTrx2 (-WCGPC-) was the same as that of PfTrx1, and different from that of PfTrx2 (-WCQAC-). The results of sequence alignments showed that BmTrx2 amino acid sequence shared a higher similarity with PfTrx1 (39%) than with PfTrx2 (26%). In the present study, we also showed that rBmTrx2 could transfer electrons to bovine insulin and be reduced by TrxR and NADPH. These results indicated that BmTrx2 might function in the regulation of redox balance and the thioredoxin systems of B. microti and P. falciparum were different from each other even though they share almost the same life-cycle during the erythrocytic stage.
In our Western blot experiments, a specific band was detected in the lysates of B. microti-infected RBCs. The molecular weight of native BmTrx2 was 15.5 kDa which was consistent with the size of the recombinant B. microti Trx2 protein. The results of the Western blot experiments showed that the expression of BmTrx2 was gradually upregulated and reached a peak at eight days post-infection. On the other hand, the results of qRT-PCR showed that the expression of BmTrx2 peaked at both three and eight days post-infection, respectively. These results are in accordance with each other. At three days post-infection, B. microti was undergoing a rapid propagation process in the host RBCs since the parasitemia increased quickly. Since it has been reported that native B. microti Trx protects DNA from oxidative damage and RBCs are oxygen-rich environments, B. microti must secrete large amounts of Trx to exert antioxidant effects [26, 27]. At a rational point of view, at three days post-infection, high expression levels of BmTrx2 helped B. microti maintain redox balance during the rapid propagation process. The B. microti parasitemia at eight days post-infection decreased to a level of 12.5% and subsequently maintained low levels. However, the level of BmTrx2 expression reached a peak at the same time. Interestingly, similar results were also observed in previous studies on other B. microti antioxidant enzymes such as peroxiredoxin 2 [14] and aldo-keto reductase-like protein [16]. This might be because of the host immune elimination. However, the molecular mechanisms of the high-level expression of BmTrx2 at eight days post-infection need to be further investigated.
Previous studies reported that P. falciparum Trx2 was located in mitochondria [28]. However, more recently, other studies revealed that PfTrx2 is part of an export machinery called the translocon of exported proteins (PTEX), which is located in the parasitophorous vacuole membrane [29, 30]. In the present study, indirect immunofluorescence assays were performed to localize BmTrx2 in B. microti merozoites. The fluorescence pattern detected suggested that the target molecules are located in the cytoplasm. However, the precise location of BmTrx2 still needs to be confirmed by co-localization with multiple markers, and molecular function of this gene needs further investigation.
In P. falciparum, PfTrx2 plays a role in the regulation of PTEX-mediated protein export and is proved to be essential in maintaining normal erythrocytic stage growth [31]. As a crucial nexus for protein export in malaria parasites, Trx2 is identified to be a potential drug target [32]. Previous studies revealed that targeting PfTrx2 pharmacologically might interfere with protein secretion, suggesting a new strategy for antimalarial drug development [33]. However apicomplexan parasites employ various mechanisms of drug resistance [34, 35], therefore, to explore the role of BmTrx2 in response to anti-babesiosis drugs and to enhance the potential of chemotherapy against babesiosis, we measured the relative expression of BmTrx2 when the parasite was subjected to chloroquine, clindamycin, quinine, and dihydroartemisinin. Our results showed that clindamycin, quinine, and dihydroartemisinin could promote BmTrx2 transcription, indicating BmTrx2 might have a role in the way these pharmaceutical agents act. However, the exact molecular mechanism of how BmTrx2 function in response to the antiprotozoal drugs still needs further exploration.
Conclusions
To our knowledge, this is the first report to identify, characterize, and functionally analyze Trx from B. microti. Our results showed that BmTrx2 is an antioxidant gene with a conserved thioredoxin domain that could be used as a potential target for the development of new drugs against babesiosis.
Abbreviations
- BmTrx2 :
-
B. microti thioredoxin 2
- DAB:
-
3, 3′-diaminobenzidine tetrahydrochloride
- Hoechst 33,342:
-
2′-[4-ethoxyphenyl]-5-[4-methyl-1-piperazinyl]-2,5′-bi-1H-benzimidazole trihydrochloride trihydrate
- IFA:
-
Immunofluorescence assay
- IPTG:
-
Isopropyl β-D-1-Thiogalactopyranoside
- LB:
-
Luria-Bertani
- PBS:
-
Phosphate-buffered saline
- qRT-PCR:
-
Quantitative real-time PCR
- RACE:
-
Rapid amplification of cDNA ends
- SDS-PAGE:
-
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- Trx:
-
Thioredoxin
- TrxR:
-
Thioredoxin reductase
References
Brennan MB, Herwaldt BL, Kazmierczak JJ, Weiss JW, Klein CL, Leith CP, et al. Transmission of Babesia microti parasites by solid organ transplantation. Emerg Infect Dis. 2016;22(11):1869.
Moniuszkomalinowska A, Swiecicka I, Dunaj J, Zajkowska J, Czupryna P, Zambrowski G, et al. Infection with Babesia microti in humans with non-specific symptoms in north-east Poland. Inf Dis. 2016;48(7):537–43.
Skrabalo Z, Deanovic Z. Piroplasmosis in man; report of a case. Doc Med Geogr Trop. 1957;9(1):11–6.
Qi C, Zhou D, Liu J, Cheng Z, Zhang L, Wang L, et al. Detection of Babesia divergens using molecular methods in anaemic patients in Shandong Province, China. Parasitol Res. 2011;109(1):241–5.
Kehr S, Sturm N, Rahlfs S, Przyborski JM, Becker K. Compartmentation of redox metabolism in malaria parasites. PLoS Pathog. 2010;6(12):e1001242.
Kawazu S, Komaki-Yasuda K, Oku H, Kano S. Peroxiredoxins in malaria parasites: parasitologic aspects. Parasitol Int. 2008;57(1):1–7.
Muller S. Redox and antioxidant systems of the malaria parasite Plasmodium falciparum. Mol Microbiol. 2004;53(5):1291–305.
Jortzik E, Becker K. Thioredoxin and glutathione systems in Plasmodium falciparum. Int J Med Microbiol. 2012;302(4–5):187–94.
Nickel C, Rahlfs S, Deponte M, Koncarevic S, Becker K. Thioredoxin networks in the malarial parasite Plasmodium falciparum. Antioxid Redox Signal. 2006;8(7–8):1227–39.
Becuwe P, Slomianny C, Valentin A, Schrevel J, Camus D, Dive D. Endogenous superoxide dismutase activity in two Babesia species. Parasitology. 1992;105(2)2:177–182.
Clarebout G, Gamain B, Precigout E, Gorenflot A, Slomianny C, Camus D, et al. Babesia hylomysci and B. divergens: presence of antioxidant enzymes destroying hydrogen peroxide. Parasitol Res. 1997;84(1):75–7.
Zhang H, Wang Z, Gong H, Cao J, Zhou Y, Zhou J. Identification and functional study of a novel 2-cys peroxiredoxin (BmTPx-1) of Babesia microti. Exp Parasitol. 2016;170:21–7.
Cornillot E, Hadj-Kaddour K, Dassouli A, Noel B, Ranwez V, Vacherie B, et al. Sequencing of the smallest apicomplexan genome from the human pathogen Babesia microti. Nucl Acids Res. 2012;40(18):9102–14.
Hai X, Zhang H, Wang Z, Gong H, Cao J, Zhou Y, et al. Identification of 2-Cys peroxiredoxin (BmTPx-2) as antioxidant active molecule from Babesia microti. Front Microbiol. 2017;8:PMC5641339.
Wang Y, Li Z, Zhou Y, Cao J, Zhang H, Gong H, et al. Specific histamine binding activity of a new lipocalin from Hyalomma asiaticum (Ixodidae) and therapeutic effects on allergic asthma in mice. Parasit Vectors. 2016;9(1):506.
Huang QCJ, Zhou Y, Huang J, Gong H, Zhang H, Zhu X-Q, Zhou J. Babesia microti aldo-keto reductase-like protein involved in antioxidant and anti-parasite response. Front Microbiol. 2017;8:2006.
Teal AE, Habura A, Ennis J, Keithly JS, Madison-Antenucci SA. New real-time PCR assay for improved detection of the parasite Babesia microti. J Clin Microbiol. 2012;50(3):903–8.
Blasa M, Angelino D, Gennari L, Ninfali P. The cellular antioxidant activity in red blood cells (CAA-RBC): a new approach to bioavailability and synergy of phytochemicals and botanical extracts. Food Chem. 2011;125(2):685–91.
Fredriksson K, Stridh H, Lundahl J, Rennard SI, Skold CM. Red blood cells inhibit proliferation and stimulate apoptosis in human lung fibroblasts in vitro. Scand J Immunol. 2004;59(6):559–65.
Zhao S, Gong H, Zhou Y, Zhang H, Cao J, Zhou J. Identification of a thioredoxin reductase from Babesia microti during mammalian infection. Parasitol Res. 2016;115(8):3219–27.
Andricopulo AD, Akoachere MB, Krogh R, Nickel C, McLeish MJ, Kenyon GL, et al. Specific inhibitors of Plasmodium falciparum thioredoxin reductase as potential antimalarial agents. Bioorg Med Chem Lett. 2006;16(8):2283–92.
Krnajski Z, Gilberger TW, Walter RD, Cowman AF, Muller S. Thioredoxin reductase is essential for the survival of Plasmodium falciparum erythrocytic stages. J Biol Chem. 2002;277(29):25970–5.
Maier AG, Rug M, O'Neill MT, Brown M, Chakravorty S, Szestak T, et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134(1):48–61.
Storm J, Perner J, Aparicio IM, Patzewitz E, Olszewski KL, Llinas M, et al. Plasmodium falciparum glutamate dehydrogenase a is dispensable and not a drug target during erythrocytic development. Malar J. 2011;10(1):193.
McCarver AC, Lessner DJ. Molecular characterization of the thioredoxin system from Methanosarcina acetivorans. FEBS J. 2014;281(20):4598–611.
Koncarevic S, Rohrbach P, Deponte M, Krohne G, Prieto JH, Yates J 3rd, et al. The malarial parasite Plasmodium falciparum imports the human protein peroxiredoxin 2 for peroxide detoxification. Proc Natl Acad Sci USA. 2009;106(32):13323–8.
Hakimi H, Goto Y, Suganuma K, Angeles JM, Kawai S, Inoue N, et al. Development of monoclonal antibodies against Plasmodium falciparum thioredoxin peroxidase 1 and its possible application for malaria diagnosis. Exp Parasitol. 2015;154:62–6.
Boucher IW, McMillan PJ, Gabrielsen M, Akerman SE, Brannigan JA, Schnick C, et al. Structural and biochemical characterization of a mitochondrial peroxiredoxin from Plasmodium falciparum. Mol Microbiol. 2006;61(4):948–59.
Kehr S, Jortzik E, Delahunty C, Yates JR 3rd, Rahlfs S, Becker K. Protein S-Glutathionylation in malaria parasites. Antioxid Redox Signal. 2011;15(11):2855–65.
De Koningward TF, Gilson PR, Boddey JA, Rug M, Smith BJ, Papenfuss AT, et al. A newly discovered protein export machine in malaria parasites. Nature. 2009;459(7249):945–9.
Matz JM, Matuschewski K, Kooij TW. Two putative protein export regulators promote Plasmodium blood stage development in vivo. Mol Biochem Parasitol. 2013;191(1):44–52.
Peng M, Cascio D, Egea PF. Crystal structure and solution characterization of the thioredoxin-2 from Plasmodium falciparum, a constituent of an essential parasitic protein export complex. Biochem Biophys Res Commun. 2015;456(1):403–9.
Sharma A, Sharma A, Dixit S, Sharma A. Structural insights into thioredoxin-2: a component of malaria parasite protein secretion machinery. Sci Rep. 2011;1:179.
Garcia-Salcedo JA, Unciti-Broceta JD, Valverde-Pozo J, Soriano M. New approaches to overcome transport related drug resistance in trypanosomatid parasites. Front Pharmacol. 2016;7:351.
White NJ. Does antimalarial mass drug administration increase or decrease the risk of resistance? Lancet Infect Dis. 2017;17(1)
Acknowledgements
We thank Dr Ibrahim Hassan from the Shanghai Veterinary Research Institute for kindly reading this manuscript and giving revision suggestions.
Funding
This work was supported by the “National Key Basic Research Program (973 program) of China” (Grant No.2015CB150300).
Availability of data and materials
All data generated or analyzed during this study are included in this article and its additional file. The sequence was submitted to the GenBank database under the accession number KX758049.1.
Author information
Authors and Affiliations
Contributions
JH and KX conceived the study, performed the statistical analyses and drafted the manuscript. YZ participated in the molecular microbiological experiments. HG and HZ participated in the design of the study and the interpretation of the data. YZ and JC participated in the experiments on animals. JZ participated in the design of the study and revision of the manuscript. JH and KX contributed equally to this work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
The protocols in this study were approved by the Institutional Animal Care and Use Committee of the Shanghai Veterinary Research Institute (IACUC approve number shvri-mo-0054), and authorized by the Animal Ethical Committee of Shanghai Veterinary Research Institute.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1: Table S1.
Detailed statistical analysis of Fig. 7. This file provides the statistical analysis details of the comparison between groups in the drug response assay. (DOCX 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Huang, J., Xiong, K., Zhang, H. et al. Molecular characterization of Babesia microti thioredoxin (BmTrx2) and its expression patterns induced by antiprotozoal drugs. Parasites Vectors 11, 38 (2018). https://doi.org/10.1186/s13071-018-2619-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-018-2619-9